Automated design of genomic Southern blot probes
Mike DR Croning, David G Fricker, Noboru H Komiyama and Seth GN Grant
Design Case - ProbeDesign39 - back to list
Summary
./analyse_probe_search_results : Mon Sep 5 11:39:14 2005
Connected to host: host, as user: user Database : southern_blot_designs Fetched probe design: ProbeDesign39 assembly : NCBIM34 bias : 3prime chromosome : X id : 15 strand : 1 ------------- Design window length: 2001bp All jobs for probe design are successful - total: 28 Fetched conf : Exonerate mouse_NCBIM34 (id: 1) Genome file(s) at: /blastdb/Mouse/NCBIM34/softmasked_dusted Fetched 557 putative probes to analyse Selection criteria to rank probes: Minimum score ratio : 10 (i.e. self-hit must score at least 10 times higher than the next best hit) Maximum percent repetitive bases: 5 Favour probes to the 3prime end of design window ------------------------ Putative probes without hits : 0 Putative probes with single_self_hit : 557/557 Putative probes exceeding selection criteria: 148/557 PASSED (GREEN) AND FAILED PUTATIVE PROBES IN THE DESIGN WINDOW --------------------------------------------------------------
Image
Unique (yellow), passed (green) and failed putative probes in the design window
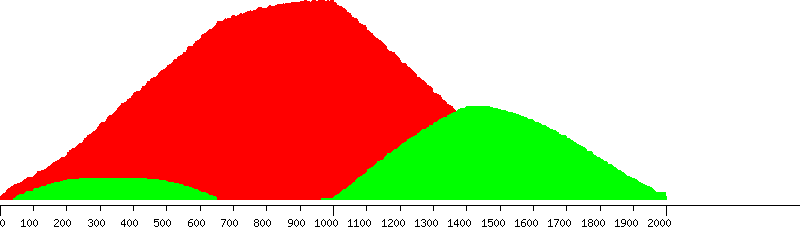
Unique putative probes
No probes
Putative probes
| Putative probe ID | Length | Score ratio | Actual | %rep DNA | Distance | Overall |
|---|---|---|---|---|---|---|
| 19112 | 950 | 29.1 | 4750 | - | 0 | 29.1 |
| 19135 | 900 | 27.6 | 4500 | - | 0 | 27.6 |
| 19136 | 900 | 27.6 | 4500 | - | 45 | 27.6 |
| 19160 | 850 | 26.1 | 4250 | - | 0 | 26.1 |
| 19161 | 850 | 26.1 | 4250 | - | 42 | 26.1 |
| 19162 | 850 | 26.1 | 4250 | - | 84 | 26.1 |
| 19188 | 800 | 24.5 | 4000 | - | 0 | 24.5 |
| 19189 | 800 | 24.5 | 4000 | - | 40 | 24.5 |
| 19190 | 800 | 24.5 | 4000 | - | 80 | 24.5 |
| 19191 | 800 | 24.5 | 4000 | - | 120 | 24.5 |
| 19192 | 800 | 24.5 | 4000 | - | 160 | 24.5 |
| 19091 | 1000 | 30.7 | 5000 | 3.2 | 0 | 24.3 |
| 19163 | 850 | 26.1 | 4250 | 0.9 | 126 | 24.2 |
| 19113 | 950 | 29.1 | 4750 | 3.1 | 47 | 23.0 |
| 19219 | 750 | 23.0 | 3750 | - | 0 | 23.0 |
| 19220 | 750 | 23.0 | 3750 | - | 37 | 23.0 |
| 19221 | 750 | 23.0 | 3750 | - | 74 | 23.0 |
| 19222 | 750 | 23.0 | 3750 | - | 111 | 23.0 |
| 19223 | 750 | 23.0 | 3750 | - | 148 | 23.0 |
| 19224 | 750 | 23.0 | 3750 | - | 185 | 23.0 |
| 19137 | 900 | 27.6 | 4500 | 2.4 | 90 | 22.7 |
| 19225 | 750 | 23.0 | 3750 | 0.5 | 222 | 21.9 |
| 19253 | 700 | 21.5 | 3500 | - | 0 | 21.5 |
| 19254 | 700 | 21.5 | 3500 | - | 35 | 21.5 |
| 19255 | 700 | 21.5 | 3500 | - | 70 | 21.5 |
| 19256 | 700 | 21.5 | 3500 | - | 105 | 21.5 |
| 19257 | 700 | 21.5 | 3500 | - | 140 | 21.5 |
| 19258 | 700 | 21.5 | 3500 | - | 175 | 21.5 |
| 19259 | 700 | 21.5 | 3500 | - | 210 | 21.5 |
| 19260 | 700 | 21.5 | 3500 | - | 245 | 21.5 |
| 19291 | 650 | 19.9 | 3250 | - | 0 | 19.9 |
| 19292 | 650 | 19.9 | 3250 | - | 32 | 19.9 |
| 19293 | 650 | 19.9 | 3250 | - | 64 | 19.9 |
| 19294 | 650 | 19.9 | 3250 | - | 96 | 19.9 |
| 19295 | 650 | 19.9 | 3250 | - | 128 | 19.9 |
| 19296 | 650 | 19.9 | 3250 | - | 160 | 19.9 |
| 19297 | 650 | 19.9 | 3250 | - | 192 | 19.9 |
| 19298 | 650 | 19.9 | 3250 | - | 224 | 19.9 |
| 19299 | 650 | 19.9 | 3250 | - | 256 | 19.9 |
| 19300 | 650 | 19.9 | 3250 | - | 288 | 19.9 |
| 19301 | 650 | 19.9 | 3250 | 0.3 | 320 | 19.3 |
| 19335 | 600 | 18.4 | 3000 | - | 30 | 18.4 |
| 19336 | 600 | 18.4 | 3000 | - | 60 | 18.4 |
| 19337 | 600 | 18.4 | 3000 | - | 90 | 18.4 |
| 19338 | 600 | 18.4 | 3000 | - | 120 | 18.4 |
| 19339 | 600 | 18.4 | 3000 | - | 150 | 18.4 |
| 19340 | 600 | 18.4 | 3000 | - | 180 | 18.4 |
| 19341 | 600 | 18.4 | 3000 | - | 210 | 18.4 |
| 19342 | 600 | 18.4 | 3000 | - | 240 | 18.4 |
| 19343 | 600 | 18.4 | 3000 | - | 270 | 18.4 |
| 19344 | 600 | 18.4 | 3000 | - | 300 | 18.4 |
| 19345 | 600 | 18.4 | 3000 | - | 330 | 18.4 |
| 19346 | 600 | 18.4 | 3000 | - | 360 | 18.4 |
| 19261 | 700 | 21.5 | 3500 | 1.7 | 280 | 18.0 |
| 19383 | 550 | 18.0 | 2750 | - | 54 | 18.0 |
| 19384 | 550 | 16.9 | 2750 | - | 81 | 16.9 |
| 19385 | 550 | 16.9 | 2750 | - | 108 | 16.9 |
| 19386 | 550 | 16.9 | 2750 | - | 135 | 16.9 |
| 19387 | 550 | 16.9 | 2750 | - | 162 | 16.9 |
| 19388 | 550 | 16.9 | 2750 | - | 189 | 16.9 |
| 19389 | 550 | 16.9 | 2750 | - | 216 | 16.9 |
| 19390 | 550 | 16.9 | 2750 | - | 243 | 16.9 |
| 19391 | 550 | 16.9 | 2750 | - | 270 | 16.9 |
| 19392 | 550 | 16.9 | 2750 | - | 297 | 16.9 |
| 19393 | 550 | 16.9 | 2750 | - | 324 | 16.9 |
| 19394 | 550 | 16.9 | 2750 | - | 351 | 16.9 |
| 19395 | 550 | 16.9 | 2750 | - | 378 | 16.9 |
| 19396 | 550 | 16.9 | 2750 | - | 405 | 16.9 |
| 19432 | 550 | 16.8 | 2750 | - | 1377 | 16.8 |
| 19440 | 500 | 15.3 | 2500 | - | 125 | 15.3 |
| 19441 | 500 | 15.3 | 2500 | - | 150 | 15.3 |
| 19442 | 500 | 15.3 | 2500 | - | 175 | 15.3 |
| 19443 | 500 | 15.3 | 2500 | - | 200 | 15.3 |
| 19444 | 500 | 15.3 | 2500 | - | 225 | 15.3 |
| 19445 | 500 | 15.3 | 2500 | - | 250 | 15.3 |
| 19446 | 500 | 15.3 | 2500 | - | 275 | 15.3 |
| 19447 | 500 | 15.3 | 2500 | - | 300 | 15.3 |
| 19448 | 500 | 15.3 | 2500 | - | 325 | 15.3 |
| 19449 | 500 | 15.3 | 2500 | - | 350 | 15.3 |
| 19450 | 500 | 15.3 | 2500 | - | 375 | 15.3 |
| 19451 | 500 | 15.3 | 2500 | - | 400 | 15.3 |
| 19452 | 500 | 15.3 | 2500 | - | 425 | 15.3 |
| 19453 | 500 | 15.3 | 2500 | - | 450 | 15.3 |
| 19490 | 500 | 15.2 | 2500 | - | 1375 | 15.2 |
| 19491 | 500 | 15.2 | 2500 | - | 1400 | 15.2 |
| 19492 | 500 | 15.2 | 2500 | - | 1425 | 15.2 |
| 19503 | 450 | 14.7 | 2250 | - | 154 | 14.7 |
| 19138 | 900 | 24.2 | 4500 | 5.0 | 135 | 14.2 |
| 19504 | 450 | 13.8 | 2250 | - | 176 | 13.8 |
| 19505 | 450 | 13.8 | 2250 | - | 198 | 13.8 |
| 19506 | 450 | 13.8 | 2250 | - | 220 | 13.8 |
| 19507 | 450 | 13.8 | 2250 | - | 242 | 13.8 |
| 19508 | 450 | 13.8 | 2250 | - | 264 | 13.8 |
| 19509 | 450 | 13.8 | 2250 | - | 286 | 13.8 |
| 19510 | 450 | 13.8 | 2250 | - | 308 | 13.8 |
| 19511 | 450 | 13.8 | 2250 | - | 330 | 13.8 |
| 19512 | 450 | 13.8 | 2250 | - | 352 | 13.8 |
| 19513 | 450 | 13.8 | 2250 | - | 374 | 13.8 |
| 19514 | 450 | 13.8 | 2250 | - | 396 | 13.8 |
| 19515 | 450 | 13.8 | 2250 | - | 418 | 13.8 |
| 19516 | 450 | 13.8 | 2250 | - | 440 | 13.8 |
| 19517 | 450 | 13.8 | 2250 | - | 462 | 13.8 |
| 19518 | 450 | 13.8 | 2250 | - | 484 | 13.8 |
| 19519 | 450 | 13.8 | 2250 | - | 506 | 13.8 |
| 19559 | 450 | 13.7 | 2250 | - | 1386 | 13.7 |
| 19560 | 450 | 13.7 | 2250 | - | 1408 | 13.7 |
| 19561 | 450 | 13.7 | 2250 | - | 1430 | 13.7 |
| 19562 | 450 | 13.7 | 2250 | - | 1452 | 13.7 |
| 19563 | 450 | 13.7 | 2250 | - | 1474 | 13.7 |
| 19433 | 550 | 16.8 | 2750 | 1.8 | 1404 | 13.1 |
| 19493 | 500 | 15.2 | 2500 | 1.2 | 1450 | 12.8 |
| 19564 | 450 | 13.7 | 2250 | 0.4 | 1496 | 12.8 |
| 19454 | 500 | 15.3 | 2500 | 1.4 | 475 | 12.5 |
| 19578 | 400 | 12.3 | 2000 | - | 220 | 12.3 |
| 19579 | 400 | 12.3 | 2000 | - | 240 | 12.3 |
| 19580 | 400 | 12.3 | 2000 | - | 260 | 12.3 |
| 19581 | 400 | 12.3 | 2000 | - | 280 | 12.3 |
| 19582 | 400 | 12.3 | 2000 | - | 300 | 12.3 |
| 19583 | 400 | 12.3 | 2000 | - | 320 | 12.3 |
| 19584 | 400 | 12.3 | 2000 | - | 340 | 12.3 |
| 19585 | 400 | 12.3 | 2000 | - | 360 | 12.3 |
| 19586 | 400 | 12.3 | 2000 | - | 380 | 12.3 |
| 19587 | 400 | 12.3 | 2000 | - | 400 | 12.3 |
| 19588 | 400 | 12.3 | 2000 | - | 420 | 12.3 |
| 19589 | 400 | 12.3 | 2000 | - | 440 | 12.3 |
| 19590 | 400 | 12.3 | 2000 | - | 460 | 12.3 |
| 19591 | 400 | 12.3 | 2000 | - | 480 | 12.3 |
| 19592 | 400 | 12.3 | 2000 | - | 500 | 12.3 |
| 19593 | 400 | 12.3 | 2000 | - | 520 | 12.3 |
| 19636 | 400 | 12.2 | 2000 | - | 1380 | 12.2 |
| 19637 | 400 | 12.2 | 2000 | - | 1400 | 12.2 |
| 19638 | 400 | 12.2 | 2000 | - | 1420 | 12.2 |
| 19639 | 400 | 12.2 | 2000 | - | 1440 | 12.2 |
| 19640 | 400 | 12.2 | 2000 | - | 1460 | 12.2 |
| 19641 | 400 | 12.2 | 2000 | - | 1480 | 12.2 |
| 19642 | 400 | 12.2 | 2000 | - | 1500 | 12.2 |
| 19643 | 400 | 12.2 | 2000 | - | 1520 | 12.2 |
| 19644 | 400 | 12.2 | 2000 | - | 1540 | 12.2 |
| 19397 | 550 | 16.9 | 2750 | 2.5 | 432 | 11.8 |
| 19558 | 450 | 13.7 | 2250 | 1.8 | 1364 | 10.2 |
| 19520 | 450 | 13.8 | 2250 | 2.2 | 528 | 9.4 |
| 19379 | 600 | 18.3 | 3000 | 4.7 | 1350 | 9.0 |
| 19431 | 550 | 16.8 | 2750 | 4.0 | 1350 | 8.8 |
| 19489 | 500 | 15.2 | 2500 | 4.4 | 1350 | 6.4 |
| 19635 | 400 | 12.2 | 2000 | 3.0 | 1360 | 6.2 |
| 19645 | 400 | 12.2 | 2000 | 4.0 | 1560 | 4.2 |
Overall score = score_ratio - 2 * repetitive DNA content
Redundant probes
| Putative probe ID | Length | Score ratio | Actual | %rep DNA | Distance | Overall |
|---|---|---|---|---|---|---|
| 19193 | 800 | 24.5 | 4000 | 4.0 | 200 | 16.5 |
| 19347 | 600 | 18.4 | 3000 | 3.7 | 390 | 11.1 |
Overall score = score_ratio - 2 * repetitive DNA content
