G2C::Proteomics
Targeted TAP purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins
Esperanza Fernandez1, Mark O Collins2, Rachel T Uren1, Maksym V Kopanitsa1, Noboru H Komiyama1, Mike DR Croning1, Lysimachos Zografos3, J Douglas Armstrong3, Jyoti S Choudhary2 and Seth GN Grant1
Author email: sg3@sanger.ac.uk * - These authors contributed equally to this work
- Genes to Cognition Programme, The Wellcome Trust Sanger Institute, Cambridge, UK
- Proteomic Mass Spectrometry, The Wellcome Trust Sanger Institute, Cambridge, UK
- School of Informatics, University of Edinburgh, Edinburgh, UK
The synapse is the connection between nerve cells (neurons). The function of the synapse is to transmit the electrical activity from one neuron to the next thus passing information through the nervous system. Inside synapses there are over a thousand proteins and they are the components from which the neurotransmission machinery is built. The organisation of these hundreds of proteins is an important issue since proteins bind and interact to form multiprotein complexes, which are molecular machines. Understanding the composition of these multiprotein complexes may shed light on diseases as well as fundamental biology of learning and other forms of behaviour.
Isolating protein complexes from cells and tissues is technically demanding and particularly from the brain, which is a very complex organ. New methods are required for complex tissues, and in this study we have further developed a system employed in yeast. Our objective was to isolate the core components of the synapse, particularly those involving proteins bound to PSD-95, which is a key molecule required for learning and memory.
We combined proteomic and mouse genetic engineering methods to isolate multiprotein complexes from the mouse brain. A Tandem Affinity Purification tag was integrated into the genomic locus of PSD-95 so that PSD-95 could be readily isolated with other proteins attached to it. This method combined with mass spectrometry analysis describes a core complex of 118 proteins that comprise containing key proteins involved in the neurotransmission.
Protein interaction network of PSD-95 interacting proteins.
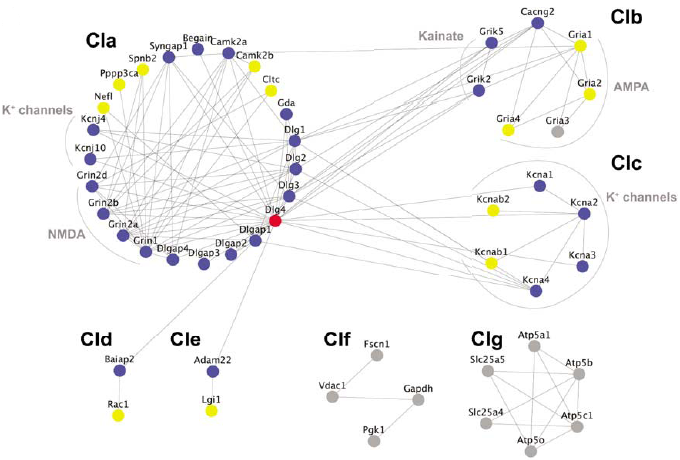
PSD-95/Dlg4 is shown in red, it's primary interactors are shown in blue and secondary interactors are shown in yellow. Click on circles to view genetic and genomic information in G2Cdb.
The medical importance of this complex is revealed since 49 proteins are involved with a range of important common brain diseases including epilepsy, depression, schizophrenia, bipolar disease, mental retardation and neurodegenerative diseases including Alzheimer's and Huntington's diseases.
Details of the mice generation and protein purification conditions are described in Fernandez et. al. The principal datasets for the synapses complex(es) are found in Table 1 and Table 2. These datasets are also integrated in Supplementary Table 1 with links to G2Cdb resources as well as the comparison with other proteomic datasets. G2Cdb access enables one to identify the functional roles for individual proteins/genes in knockout mice and human genetics.
Data resources
- Table 1. Functional classification of PSD-95-associated proteins in at least three out of four tandem purifications.
- Table 2. Functional classification of PSD-95-associated proteins in one or two tandem purifications.
- Table 3. Genes associated with neurological and psychiatric diseases.
- Supplementary Table 1. Proteins identified in the single step and tandem purifications by LC-MS/MS.
- Supplementary Table 2. Proteins identified by LC-MS/MS in three tandem purifications from wild type mice.
- Supplementary Table 3. Known PSD-95 primary interactors extracted from UniHI database and manually curated.
- Supplementary Table 4. EmPAI values from single step and tandem purifications.
- Supplementary Table 5. Comparison to proteins found in receptor complexes and synaptic lists.
- Supplementary Table 6. Protein nodes of the interaction network's MCC.
- Supplementary Table 7. Gene targeting and genotyping primers.
FAQs
1. What is the Tandem Affinity Purification (TAP) method?
The Tandem Affinity Purification method consists of a genetic modification of the protein of interest by fusion with a TAP (two different tags separated by a protease site) tag into the C- or N- terminal end of the protein (10504710). The tagged protein is isolated (with its associated proteins) in a tandem procedure: a first purification is followed by a protease cleavage and the complex is recovered in a second purification that ends with the elution in native conditions.
2. What advantages does the TAP method offer?
The TAP method allows a more stringent purification removing weak interactors and potential contaminants. The TAP method also has technical advantages over immunoprecipitation that is limited by i) the availability of suitable antibodies and their cross reaction with other proteins, ii) the possibility that the antibody-protein interaction is affected by either posttranslational modifications or by the binding with other proteins, iii) the antibody may disrupt interacting partners, and iv) the harsh conditions for the complex elution may result in protein degradation.
3. Which proteins belong to the core set of postsynaptic proteins?
Table 1 lists the proteins identified in at least three of four Tandem Affinity Purifications that are considered as the core of the postsynaptic terminal of excitatory synapses. Table 2 shows the synaptic proteins that have been purified once or twice and could be weak or non-abundant PSD-95 interactors. These proteins should not be considered as part of the PSD-95 core complex.
4. Has this protein been identified in any PSD purification and/or in the MASC complex, PSD or other neuronal complex before?
Go to Supplementary Table 1 and identify the gene name or the protein ID. Follow the G2Cdb link and the G2Cdb gene report is shown. Other gene lists containing this gene are reported.
5. Does this protein directly bind PSD-95?
PSD-95 binary interactors that belong to the PSD-95 complex are listed in Supplementary Table 3. Literature reference are provided directly to PubMed for inspection.
6. What is the G2C database?
G2Cdb is an integrated database for neurobiologists that contains data linking gene function to cognition. The datasets include gene and protein expression, synaptic physiology, mouse and human behaviour data with synapse proteomics. The datasets includes that generated by members of the Genes to Cognition program and literature mined data.
7. Is this gene related to any disorder?
PSD-95 interactors involved in mental disease are shown in the Table 3. Following the G2Cdb ID in the Supplementary Table 1 you have access to the G2Cdb gene report that links to behavioural and physiological data in knockout mice.
References
Table 1. Functional classification of PSD-95-associated proteins in at least three out of four tandem purifications.
Approved gene symbols and protein names, and UniProt accession numbers are shown. Number of approved peptides for each protein identified by LC-MS/MS in the four tandem purifications are indicated as T1, T2, T3 and T4. More information of these proteins is listed in Supplementary Table 1.
Adaptor / Regulatory Proteins
| Number of peptides | |||||||
|---|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 | Cluster |
| Anks1 | ankyrin repeat and SAM domain containing 1 | P59672 | 4 | 3 | 3 | ||
| Anks1b | ankyrin repeat and sterile alpha motif domain containing 1B | Q8BZM2 | 17 | 5 | 10 | 10 | |
| Baiap2 | brain-specific angiogenesis inhibitor 1-associated protein 2 | Q8BKX1 | 20 | 12 | 13 | 13 | d |
| Begain | brain-enriched guanylate kinase-associated | Q68EF6 | 4 | 8 | 12 | 16 | a |
| Dlg1 | Synapse-associated protein 97 | Q3UP61 | 24 | 42 | 34 | 42 | a |
| Dlg2 | Postsynaptic density protein 93 | Q91XM9 | 49 | 67 | 69 | 80 | a |
| Dlg3 | Synapse-associated protein 102 | Q52KF7 | 27 | 17 | 14 | 22 | a |
| Dlg4 | Postsynaptic density protein 95 | Q62108 | 42 | 57 | 64 | 64 | a |
| Dlgap1 | SAP90/PSD-95-associated protein 1 | Q9D415 | 11 | 9 | 14 | 24 | a |
| Dlgap2 | SAP90/PSD-95-associated protein 2 | Q8BJ42 | 18 | 12 | 19 | 24 | a |
| Dlgap3 | SAP90/PSD-95-associated protein 3 | A2A7T7 | 16 | 8 | 7 | 15 | a |
| Dlgap4 | SAP90/PSD-95-associated protein 4 | A2BDU3 | 14 | 9 | 7 | 14 | a |
Receptors / Channels / Transporters Proteins
| Number of peptides | |||||||
|---|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 | Cluster |
| Gria1 | glutamate receptor, ionotropic, AMPA 1 | Q5NBY1 | 2 | 3 | 5 | 13 | b |
| Gria2 | glutamate receptor, ionotropic, AMPA 2 | P23819 | 4 | 8 | 10 | 20 | b |
| Gria3 | glutamate receptor, ionotrophic, AMPA 3 | Q9Z2W9 | 2 | 9 | 18 | b | |
| Gria4 | glutamate receptor, ionotrophic, AMPA 4 | Q9Z2W8 | 2 | 2 | 5 | 9 | b |
| Grik2 | glutamate receptor, ionotropic, kainate 2 (beta 2) | P39087 | 2 | 4 | 6 | b | |
| Grik5 | glutamate receptor, ionotropic, kainate 5 (gamma 2) | Q61626 | 2 | 3 | 7 | b | |
| Grin1 | glutamate receptor, ionotropic, NMDA1 (zeta 1) | A2AI21 | 29 | 40 | 50 | 55 | a |
| Grin2a | glutamate receptor, ionotropic, NMDA2A (epsilon 1) | P35436 | 24 | 31 | 36 | 46 | a |
| Grin2b | glutamate receptor, ionotropic, NMDA2B (epsilon 2) | Q01097 | 44 | 54 | 67 | 78 | a |
| Grin2d | glutamate receptor, ionotropic, NMDA2D (epsilon 4) | Q03391 | 3 | 6 | 9 | 10 | a |
| Gpr123 | G protein-coupled receptor 123 | Q52KJ6 | 2 | 3 | 3 | 3 | |
| Cacng2 | calcium channel, voltage-dependent, gamma subunit 2 | O88602 | 2 | 2 | 2 | 3 | b |
| Kcna1 | K+ voltage-gated channel, shaker-related subfamily, member 1 | P16388 | 6 | 5 | 5 | 6 | c |
| Kcna2 | K+ voltage-gated channel, shaker-related subfamily, member 2 | P63141 | 4 | 5 | 7 | 6 | c |
| Kcna3 | K+ voltage-gated channel, shaker-related subfamily, member 3 | P16390 | 3 | 4 | 6 | 5 | c |
| Kcna4 | K+ voltage-gated channel, shaker-related subfamily, member 4 | Q8CBF8 | 2 | 3 | 5 | 5 | c |
| Kcnab1 | K+ voltage-gated channel, shaker-related subfamily, beta member 1 | P63143 | 3 | 3 | 4 | 6 | c |
| Kcnab2 | K+ voltage-gated channel, shaker-related subfamily, beta member 2 | P62482 | 5 | 6 | 10 | 11 | c |
| Kcnj10 | K+ inwardly-rectifying channel, subfamily J, member 10 | Q9JM63 | 3 | 3 | 4 | 6 | a |
| Kcnj4 | K+ inwardly-rectifying channel, subfamily J, member 4 | P52189 | 4 | 6 | 6 | 8 | a |
| Vdac1 | voltage-dependent anion channel 1 | Q60932 | 4 | 4 | 5 | 5 | f |
| Vdac2 | voltage-dependent anion channel 2 | Q60930 | 4 | 2 | 3 | 4 | |
| Atp1b1 | ATPase, Na+/K+ transporting, beta 1 polypeptide1 | P14094 | 3 | 3 | 6 | 3 | |
| Atp6v0d1 | ATPase, H+ transporting, V0 subunit d isoform 1 | P51863 | 3 | 3 | 4 | 2 | |
| Sfxn3 | sideroflexin 3 | Q91V61 | 3 | 3 | 4 | 3 | |
| Slc1a2 | solute carrier family, member 2 | P43006 | 3 | 2 | 4 | 4 | |
| Slc25a4 | ADP/ATP translocase 1 | P48962 | 2 | 3 | 6 | g | |
| Slc25a5 | ADP/ATP translocase 2 | P51881 | 2 | 2 | 3 | 6 | g |
| Slc4a4 | solute carrier family 4 (anion exchanger), member 4 | O88343 | 3 | 6 | 4 | ||
Cytoskeletal / Structural / Cell adhesion Proteins
| Number of peptides | |||||||
|---|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 | Cluster |
| Ablim1 | actin binding LIM protein 1 | Q8K4G5 | 20 | 10 | 18 | 22 | |
| Adam22 | a disintegrin and metallopeptidase domain 22 | Q9R1V6 | 12 | 16 | 24 | 31 | e |
| Arc | activity regulated cytoskeletal-associated protein | Q9WV31 | 8 | 13 | 20 | 19 | |
| Arpc4 | actin related protein 2/3 complex, subunit 4 | P59999 | 2 | 2 | 4 | ||
| Capza2 | capping protein (actin filament) muscle Z-line, alpha 2 | P47754 | 2 | 2 | 2 | ||
| Cfl1 | cofilin 1, non-muscle | P18760 | 2 | 2 | 3 | 4 | |
| Dstn | destrin | Q9R0P5 | 2 | 5 | 6 | 6 | |
| Fscn1 | fascin homolog 1, actin bundling protein (Strongylocentrotus purpuratus) | Q61553 | 2 | 2 | 3 | f | |
| Lgi1 | leucine-rich repeat LGI family, member 1 | Q9JIA1 | 6 | 12 | 19 | 15 | e |
| Nefl | neurofilament, light polypeptide 68kDa | P08551 | 3 | 3 | 6 | a | |
| Nrxn1 | neurexin 1 | Q9CS84 | 8 | 17 | 12 | 23 | |
| Plp1 | proteolipid protein (myelin) 1 | P60202 | 2 | 3 | 4 | 5 | |
| Sept11 | septin 11 | Q8C1B7 | 2 | 2 | 2 | 2 | |
| Sept5 | septin 5 | Q9Z2Q6 | 2 | 2 | 4 | ||
| Spnb2 | spectrin beta 2 | Q62261 | 2 | 2 | 5 | a | |
| Tuba1a* | tubulin, beta polypeptide | P05213, | 11 | 14 | 20 | 20 | |
| Tubb2b* | tubulin, beta 2b | Q7TMM9 | 19 | 21 | 24 | 27 | |
| Tubb6 | tubulin, beta 6 | Q922F4 | 8 | 8 | 13 | 13 | |
Vesicular / Trafficking / Transport Proteins
| Number of peptides | |||||||
|---|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 | Cluster |
| Arf3 | ADP-ribosylation factor 3 | P61205 | 2 | 2 | 3 | 4 | |
| Cltc | clathrin, heavy chain (Hc) | Q5SXR6 | 10 | 4 | 12 | a | |
| Cpne4* | copine IV | Q8BLR2 | 4 | 2 | 3 | ||
| Cpne7 | copine VII7 | Q0VE82 | 4 | 3 | 2 | 3 | |
| Iqsec1 | IQ motif and Sec7 domain 1 | Q8R0S2 | 11 | 7 | 6 | 15 | |
| Iqsec2 | IQ motif and Sec7 domain 2 | Q5DU25 | 34 | 25 | 24 | 36 | |
| Nsf | N-ethylmaleimide sensitive fusion protein | P46460 | 4 | 4 | 7 | 9 | |
| Stx1b2 | syntaxin 1B2 | P61264 | 3 | 4 | 3 | 2 | |
| Stxbp1 | syntaxin binding protein 1 | O08599 | 5 | 3 | 4 | 4 | |
| Syt1 | synaptotagmin I | P46096 | 5 | 2 | 2 | 2 | |
| Vamp2* | Synaptobrevin 2 | P63024 | 2 | 4 | 2 | ||
Enzyme Proteins
| Number of peptides | |||||||
|---|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 | Cluster |
| Acat1 | acetyl-Coenzyme A acetyltransferase 1 | Q8QZT1 | 2 | 3 | 3 | ||
| Aco2 | aconitase 2, mitochondrial | Q99KI0 | 3 | 3 | 2 | ||
| Acot7 | acyl-CoA thioesterase 7 | Q91V12 | 3 | 3 | 3 | 4 | |
| Aldoc | aldolase C, fructose-bisphosphate | P05063 | 4 | 2 | 4 | ||
| Atp5c1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | Q91VR2 | 3 | 3 | 4 | g | |
| Atp5b | ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit | P56480 | 2 | 6 | 5 | g | |
| Atp5o | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | Q9DB20 | 2 | 2 | 4 | 4 | g |
| Atp5a1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 1 | Q03265 | 5 | 10 | 14 | 11 | g |
| Cnp | 2',3'-cyclic nucleotide 3' phosphodiesterase | P16330 | 10 | 3 | 11 | 10 | |
| Gapdh | glyceraldehyde-3-phosphate dehydrogenase | P16858 | 9 | 8 | 9 | 10 | f |
| Gda | guanine deaminase | Q9R111 | 29 | 26 | 29 | 31 | a |
| Glul | glutamate-ammonia ligase (glutamine synthetase) | P15105 | 5 | 17 | 21 | 19 | |
| Gpx4 | glutathione peroxidase 4 | O70325 | 2 | 2 | 2 | ||
| Msrb2 | methionine sulfoxide reductase B2 | Q78J03 | 2 | 7 | 4 | ||
| Pdha1 | pyruvate dehydrogenase E1 alpha 1 | P35486 | 4 | 3 | 8 | 5 | |
| Pdhb | pyruvate dehydrogenase (lipoamide) beta | Q9D051 | 4 | 4 | 4 | 6 | |
| Pgk1 | phosphoglycerate kinase 1 | P09411 | 2 | 3 | 13 | 8 | f |
| Pkm2 | pyruvate kinase, muscle | P52480 | 7 | 6 | 11 | 12 | |
| Ppap2b | phosphatidic acid phosphatase type 2B | Q99JY8 | 2 | 2 | 5 | ||
| Prdx1 | peroxiredoxin 1 | P35700 | 4 | 7 | 10 | 14 | |
| Prdx2 | peroxiredoxin 2 | Q61171 | 2 | 2 | 3 | ||
| Sdha | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | Q8K2B3 | 7 | 6 | 8 | 6 | |
| Sucla2 | succinate-CoA ligase, ADP-forming, beta subunit | Q9Z2I9 | 4 | 2 | 5 | 6 | |
Kinase Proteins
| Number of peptides | |||||||
|---|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 | Cluster |
| Camk2a | calcium/calmodulin-dependent protein kinase II alpha | P11798 | 11 | 3 | 4 | 11 | a |
| Camk2b | calcium/calmodulin-dependent protein kinase II beta | Q5SVI3 | 10 | 5 | 5 | 7 | a |
| Mapk1 | mitogen activated protein kinase 1 | P63085 | 3 | 3 | 4 | 9 | |
| Phosphatases | |||||||
| Ppap2b | phosphatidic acid phosphatase type 2B | Q99JY8 | 2 | 2 | 5 | ||
| Ppp3ca | protein phosphatase 3, catalytic subunit, alpha isoform | P63328 | 12 | 6 | 7 | 11 | a |
| Ppp3cb | protein phosphatase 3, catalytic subunit, beta isoform | P48453 | 7 | 3 | 2 | 4 | |
G-protein signaling Proteins
| Number of peptides | |||||||
|---|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 | Cluster |
| Abr | active BCR-related gene | Q6PCY1 | 2 | 2 | 6 | ||
| Gnao1 | guanine nucleotide binding protein, alpha o | P18872 | 8 | 7 | 15 | 16 | |
| Kalrn | kalirin, RhoGEF kinase | A2CG52 | 2 | 3 | 2 | 8 | |
| Rac1 | RAS-related C3 botulinum substrate 1 | Q3TLP8 | 2 | 2 | 3 | 3 | d |
| Syngap1 | synaptic Ras GTPase activating protein 1 homolog (rat) | Q9QUH6 | 21 | 15 | 17 | 38 | a |
Transcription / Translation Proteins
| Number of peptides | |||||||
|---|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 | Cluster |
| Park7 | Parkinson disease (autosomal recessive, early onset) 7 | A2A817 | 3 | 3 | 3 | ||
| Rps14 | ribosomal protein S14 | P62264 | 2 | 3 | 2 | ||
| Rps3 | ribosomal protein S3 | P62908 | 2 | 3 | 4 | ||
| Uba52* | ubiquitin A-52 residue ribosomal protein fusion product 1 | Q66JP1 | 3 | 3 | 5 | 6 | |
Signal transduction Proteins
| Number of peptides | |||||||
|---|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 | Cluster |
| Btbd11 | BTB (POZ) domain containing 11 | Q6GQW0 | 5 | 3 | 2 | 3 | |
| Phb2 | prohibitin 2 | O35129 | 2 | 5 | 4 | ||
| Ywhae | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | P62259 | 3 | 2 | 2 | ||
| Pcbp1 | poly(rC) binding protein 1 | P60335 | 3 | 3 | 5 | ||
Unclassified Proteins
| Number of peptides | |||||||
|---|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 | Cluster |
| Fam81a | family with sequence similarity 81, member A | Q3UXZ6 | 7 | 6 | 21 | 18 | |
| AI662250 | expressed sequence AI662250 | Q3UKV2 | 2 | 2 | 2 | ||
| B630019K06Rik | RIKEN cDNA B630019K06 gene | Q7TNS5 | 6 | 7 | 8 | 9 | |
| Frmpd3 | FERM and PDZ domain containing 3 | Q8BXG0 | 2 | 5 | 5 | ||
| Pgam5 | phosphoglycerate mutase family member 5 | Q3UK19 | 7 | 7 | 8 | 10 | |
| Prrt1 | proline-rich transmembrane protein 1 | O35449 | 2 | 2 | 2 | 3 | |
| Slc9a3r1 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | P70441 | 4 | 3 | 7 | 2 | |
Genes marked with an asterisk represent genes whose peptides are common to other genes:
- Cpne5*
- Cpne4, Cpne8.
- Tuba1a*
- Tuba1b, Tuba4c, Tuba1b.
- Tubb2b*
- Tubb5, Tubb2a, Tubb2c, Tubb4.
- Uba52*
- Ubc, Ubb.
- Vamp2*
- Vamp3.
Table 2. Functional classification of PSD-95-associated proteins in one or two tandem purifications.
Adaptor / Regulatory Proteins
| Number of peptides | ||||||
|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 |
| Ap2a1 | adaptor protein complex AP-2, alpha 1 subunit | P17426 | 3 | |||
| Grb2 | growth factor receptor bound protein 2 | Q60631 | 2 | 4 | ||
Receptors / Channels / Transporters Proteins
| Number of peptides | ||||||
|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 |
| Atp2b1 | ATPase, Ca++ transporting, plasma membrane 1 | Q05CJ5 | 2 | 4 | ||
| Grin2c | glutamate receptor, ionotropic, NMDA2C (epsilon 3) | Q01098 | 5 | |||
| Grm3 | glutamate receptor, metabotropic 3 | Q9QYS2 | 2 | |||
| Kcnj16 | potassium inwardly-rectifying channel, subfamily J, member 16 | Q9Z307 | 7 | |||
| Lrp1 | low density lipoprotein receptor-related protein 1 | Q91ZX7 | 2 | 3 | ||
| Lrrtm1 | leucine rich repeat transmembrane neuronal 1 | Q8K377 | 2 | |||
| Slc1a3 | Sodium-dependent glutamate/aspartate transporter 1 | P56564 | 3 | 3 | ||
| Slc2a1 | solute carrier family 2 (facilitated glucose transporter), member 1 | P17809 | 2 | 2 | ||
| Vdac3 | voltage-dependent anion channel 3 | Q60931 | 2 | 5 | ||
Cytoskeletal / Structural / Cell adhesion Proteins
| Number of peptides | ||||||
|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 |
| Mtap1a | microtubule-associated protein 1 A | Q9QYR6 | 3 | 2 | ||
| Nlgn2 | neuroligin 2 | Q69ZK9 | 2 | |||
| Nlgn3 | neuroligin 3 | A2AGI2 | 2 | |||
| Syn2 | synapsin II | Q64332 | 3 | 4 | ||
| Shank1 | SH3/ankyrin domain gene 1 | XP_001474960 | 4 | |||
Enzyme Proteins
| Number of peptides | ||||||
|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 |
| Cit | citron | P49025 | 11 | |||
| Crym | crystallin, mu | O54983 | 3 | 3 | ||
| Csmd2 | CUB and Sushi multiple domains 2 | A2A8D7 | 5 | |||
| Dusp10 | dual specificity phosphatase 10 | Q8R3L3 | 3 | 2 | ||
| Jak3 | Janus kinase 3 | Q62137 | 2 | |||
| Mapk3 | mitogen activated protein kinase 3 | Q63844 | 4 | |||
| Ube2v1 | ubiquitin-conjugating enzyme E2 variant 1 | Q9CZY3 | 2 | 5 | ||
| Ube2v2 | ubiquitin-conjugating enzyme E2 variant 2 | Q9D2M8 | 2 | 4 | ||
G-protein signaling Proteins
| Number of peptides | ||||||
|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 |
| Gna13 | guanine nucleotide binding protein, alpha 13 | Q8C5L2 | 2 | 2 | ||
| Gnb1 | guanine nucleotide binding protein (G protein), beta 1 | P62874 | 3 | 3 | ||
| Rab6 | RAB6, member RAS oncogene family | P35279 | 4 | |||
Signalling Proteins
| Number of peptides | ||||||
|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 |
| Fbxo2 | F-box protein 2 | Q80UW2 | 4 | |||
| Fbxo6 | F-box protein 6 | Q9QZN4 | 3 | 4 | ||
| Nxph3 | neurexophilin 3 | Q91VX5 | 2 | |||
| Pcbp2 | poly(rC) binding protein 2 | Q61990 | 3 | 3 | ||
| Traf3 | Tnf receptor-associated factor 3 | Q3UHJ1 | 3 | 3 | ||
Chaperone / Protein folding / Signalling Proteins
| Number of peptides | ||||||
|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 |
| Hspa12a | heat shock protein 12A | Q8K0U4 | 2 | |||
| DNA binding | ||||||
| Hist1h2bj* | histone 1, H2bb | Q8CGP2 | 3 | 3 | ||
Transcription / Translation Proteins
| Number of peptides | ||||||
|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 |
| Eef1a1* | eukaryotic translation elongation factor 1 alpha 1 | P10126 | 2 | 2 | ||
| Lsm11 | U7 snRNP-specific Sm-like protein LSM11 | Q8BUV6 | 2 | 3 | ||
Unclassified Proteins
| Number of peptides | ||||||
|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | UniProt Acc | T1 | T2 | T3 | T4 |
| Clu | clusterin | Q06890 | 2 | 3 | ||
| Lancl1 | LanC (bacterial lantibiotic synthetase component C)-like 1 | O89112 | 4 | |||
| Mog | myelin oligodendrocyte glycoprotein | Q61885 | 2 | 2 | ||
| Neto1 | neuropilin (NRP) and tolloid (TLL)-like 1 | Q8R4I7 | 2 | 2 | ||
+ indicates NCBI accession number.
Genes marked with an asterisk represent genes whose peptides are common to other genes:
- Hist1h2bj*
- Hist1h2bm Hist1h2be Hist1h2bn Hist1h2bg Hist1h2bp Hist1h2bh Hist1h2bf Hist1h2bb Hist3h2bb Hist1h2bc Hist1h2bl Hist2h2bb.
Table 3. Genes associated with neurological and psychiatric diseases.
Disease association data for proteins in the tandem purification was mined from the Genetic Association Database, CiteXplore and manually curated. References are provided below.
- Adam22
- Epilepsy1
- Acot7
- Schizophrenia3
- Atp1b1
- Rett syndrome5
- Neurodegeneration7
- Atp5a1
- Alzheimer's9
- Atp5c1
- Bipolar afective disorder11, 12
- Cacng2
- Bipolar disorder13
- CamKIIa
- Bipolar disorder14
- CamKIIb
- Schizophrenia17
- Depression17
- Capza2
- Mental retardation19
- Cltc
- Mental retardation21
- Cnp1
- Schizophrenia3, 22
- Dlg1
- Schizophrenia24
- Dlg2
- Schizophrenia22
- Dlg3
- Schizophrenia23, 24
- Bipolar disorder23
- Depression23
- X-Mental retardation31
- Dlg4
- Schizophrenia33
- Bipolar disorder23
- Dlgap1
- Schizophrenia35
- Gapdh
- Alzheimer's39
- Gda
- Schizophrenia37
- Gnao1
- Schizophrenia41
- Gria1
- Schizophrenia46-50
- Alzheimer's55-59
- Epilepsy61, 62
- Gria2
- Schizophrenia4, 64
- Epilepsy66-71
- Gria3
- Schizophrenia47
- X-Mental retardation73
- Gria4
- Schizophrenia22
- Grik2
- Mental retardation75
- Schizophrenia22
- Grin1
- Attention disorder78
- Bipolar afective disorder80
- Schizophrenia81
- Seizure82
- Grin2a
- Alzheimer's83
- Huntington disease84
- Schizophrenia33
- Grin2b
- Schizophrenia22, 23
- Bipolar afective disorder86-88
- Epilepsy11, 89
- Huntington disease84, 90
- Grin2d
- Schizophrenia2
- Kcnj4
- Schizophrenia4
- Kcna1
- Episodic ataxia, type 16
- Kcnj10
- Epilepsy8
- Seizure10
- Lgi1
- Epilepsy1
- Mapk1
- Schizophrenia15, 16
- Depression18
- Msrb2
- Bipolar afective disorder11
- Nefl
- CMT120
- Schizophrenia3
- Bipolar23
- CMT225, 26
- ALS27, 28
- Nrxn1
- Autism29
- Schizophrenia22
- Nsf
- Schizophrenia30
- Pdha1
- Depression32
- Pgk1
- Parkinson's5
- Mental retardation34
- Bipolar disorder36
- Plp1
- Pelizaeus-Merzbacher disease38
- Depression40
- Multiple sclerosis42-45
- Demyelinating disease51-54
- Spastic paraplegia60
- Pppp3ca
- Schizophrenia63
- Prdx1
- Alzheimer's65
- Prdx2
- Parkinson's72
- Sl1a2
- Schizophrenia22
- ALS74
- Slc25a4
- Bipolar afective disorder12
- Ophtalmoplegia76
- Stxbp1
- Schizophrenia77
- Vdac1
- Alzheimer's9
- Schizophrenia3
- Bipolar afective disorder12
- Vdac2
- Bipolar afective disorder12
- Ywhae
- Miller-Dieker lissencephaly85
References
- Fukata, Y. et al. Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science 313, 1792-5 (2006).
- Makino, C., Shibata, H., Ninomiya, H., Tashiro, N. & Fukumaki, Y. Identification of single-nucleotide polymorphisms in the human N-methyl-D-aspartate receptor subunit NR2D gene, GRIN2D, and association study with schizophrenia. Psychiatr Genet 15, 215-21 (2005).
- Hakak, Y. et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci U S A 98, 4746-51 (2001).
- Le-Niculescu, H. et al. Towards understanding the schizophrenia code: an expanded convergent functional genomics approach. Am J Med Genet B Neuropsychiatr Genet 144B, 129-58 (2007).
- Delgado, I. J., Kim, D. S., Thatcher, K. N., LaSalle, J. M. & Van den Veyver, I. B. Expression profiling of clonal lymphocyte cell cultures from Rett syndrome patients. BMC Med Genet 7, 61 (2006).
- Browne, D. L. et al. Identification of two new KCNA1 mutations in episodic ataxia/myokymia families. Hum Mol Genet 4, 1671-2 (1995).
- Weber, P., Bartsch, U., Schachner, M. & Montag, D. Na,K-ATPase subunit beta1 knock-in prevents lethality of beta2 deficiency in mice. J Neurosci 18, 9192-203 (1998).
- Lenzen, K. P. et al. Supportive evidence for an allelic association of the human KCNJ10 potassium channel gene with idiopathic generalized epilepsy. Epilepsy Res 63, 113-8 (2005).
- Zhang, J. et al. Inhibition of the dopamine D1 receptor signaling by PSD-95. J Biol Chem 282, 15778-89 (2007).
- Buono, R. J. et al. Association between variation in the human KCNJ10 potassium ion channel gene and seizure susceptibility. Epilepsy Res 58, 175-83 (2004).
- Conti, L. et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol 3, e283 (2005).
- Konradi, C. et al. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry 61, 300-8 (2004).
- Silberberg, G. et al. Stargazin involvement with bipolar disorder and response to lithium treatment. Pharmacogenet Genomics 18, 403-12 (2008).
- Xing, G. et al. Decreased prefrontal CaMKII alpha mRNA in bipolar illness. Neuroreport 13, 501-5 (2002).
- Kyosseva, S. V. Differential expression of mitogen-activated protein kinases and immediate early genes fos and jun in thalamus in schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry 28, 997-1006 (2004).
- Kyosseva, S. V. et al. Mitogen-activated protein kinases in schizophrenia. Biol Psychiatry 46, 689-96 (1999).
- Novak, G., Seeman, P. & Tallerico, T. Increased expression of calcium/calmodulindependent protein kinase IIbeta in frontal cortex in schizophrenia and depression. Synapse 59, 61-8 (2006).
- Dwivedi, Y. et al. Reduced activation and expression of ERK1/2 MAP kinase in the post-mortem brain of depressed suicide subjects. J Neurochem 77, 916-28 (2001).
- Gulesserian, T., Kim, S. H., Fountoulakis, M. & Lubec, G. Aberrant expression of centractin and capping proteins, integral constituents of the dynactin complex, in fetal down syndrome brain. Biochem Biophys Res Commun 291, 62-7 (2002).
- Jordanova, A. et al. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot-Marie-Tooth disease. Brain 126, 590-7 (2003).
- Holmes, S. E. et al. Disruption of the clathrin heavy chain-like gene (CLTCL) associated with features of DGS/VCFS: a balanced (21;22)(p12;q11) translocation. Hum Mol Genet 6, 357-67 (1997).
- Carter, C. J. eIF2B and oligodendrocyte survival: where nature and nurture meet in bipolar disorder and schizophrenia? Schizophr Bull 33, 1343-53 (2007).
- Clinton, S. M. & Meador-Woodruff, J. H. Abnormalities of the NMDA Receptor and Associated Intracellular Molecules in the Thalamus in Schizophrenia and Bipolar Disorder. Neuropsychopharmacology 29, 1353-62 (2004).
- Toyooka, K. et al. Selective reduction of a PDZ protein, SAP-97, in the prefrontal cortex of patients with chronic schizophrenia. J Neurochem 83, 797-806 (2002).
- Georgiou, D. M. et al. A novel NF-L mutation Pro22Ser is associated with CMT2 in a large Slovenian family. Neurogenetics 4, 93-6 (2002).
- Miltenberger-Miltenyi, G. et al. Clinical and electrophysiological features in Charcot- Marie-Tooth disease with mutations in the NEFL gene. Arch Neurol 64, 966-70 (2007).
- Cluskey, S. & Ramsden, D. B. Mechanisms of neurodegeneration in amyotrophic lateral sclerosis. Mol Pathol 54, 386-92 (2001).
- Lee, I. et al. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat Genet 40, 181-8 (2008).
- Szatmari, P. et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 39, 319-28 (2007).
- Mirnics, K., Middleton, F. A., Marquez, A., Lewis, D. A. & Levitt, P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron 28, 53-67 (2000).
- Tarpey, P. et al. Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am J Hum Genet 75, 318-24 (2004).
- Koene, S. et al. Major depression in adolescent children consecutively diagnosed with mitochondrial disorder. J Affect Disord (2008).
- Dracheva, S. et al. N-methyl-D-aspartic acid receptor expression in the dorsolateral prefrontal cortex of elderly patients with schizophrenia. Am J Psychiatry 158, 1400-10 (2001).
- Svaasand, E. K., Aasly, J., Landsem, V. M. & Klungland, H. Altered expression of PGK1 in a family with phosphoglycerate kinase deficiency. Muscle Nerve 36, 679-84 (2007).
- Pickard, B. S. et al. Candidate psychiatric illness genes identified in patients with pericentric inversions of chromosome 18. Psychiatr Genet 15, 37-44 (2005).
- Matigian, N. et al. Expression profiling in monozygotic twins discordant for bipolar disorder reveals dysregulation of the WNT signalling pathway. Mol Psychiatry 12, 815-25 (2007).
- Pennington, K. et al. Prominent synaptic and metabolic abnormalities revealed by proteomic analysis of the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder. Mol Psychiatry 13, 1102-17 (2008).
- Kutsuwada, T. et al. Impairment of suckling response, trigeminal neuronal pattern formation, and hippocampal LTD in NMDA receptor epsilon 2 subunit mutant mice. Neuron 16, 333-44 (1996).
- Yao, W. D. et al. Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron 41, 625-38 (2004).
- Sokolov, B. P. Oligodendroglial abnormalities in schizophrenia, mood disorders and substance abuse. Comorbidity, shared traits, or molecular phenocopies? Int J Neuropsychopharmacol 10, 547-55 (2007).
- Vawter, M. P. et al. Microarray screening of lymphocyte gene expression differences in a multiplex schizophrenia pedigree. Schizophr Res 67, 41-52 (2004).
- Jamain, S. et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet 34, 27-9 (2003).
- Trotter, J. L., Hickey, W. F., van der Veen, R. C. & Sulze, L. Peripheral blood mononuclear cells from multiple sclerosis patients recognize myelin proteolipid protein and selected peptides. J Neuroimmunol 33, 55-62 (1991).
- Klein, L., Klugmann, M., Nave, K. A., Tuohy, V. K. & Kyewski, B. Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nat Med 6, 56-61 (2000).
- Chou, Y. K. et al. Frequency of T cells specific for myelin basic protein and myelin proteolipid protein in blood and cerebrospinal fluid in multiple sclerosis. J Neuroimmunol 38, 105-13 (1992).
- O'Connor, J. A. & Hemby, S. E. Elevated GRIA1 mRNA expression in Layer II/III and V pyramidal cells of the DLPFC in schizophrenia. Schizophr Res 97, 277-88 (2007).
- Magri, C. et al. Study on GRIA2, GRIA3 and GRIA4 genes highlights a positive association between schizophrenia and GRIA3 in female patients. Am J Med Genet B Neuropsychiatr Genet 147B, 745-53 (2008).
- Eastwood, S. L. et al. Decreased expression of mRNAs encoding non-NMDA glutamate receptors GluR1 and GluR2 in medial temporal lobe neurons in schizophrenia. Brain Res Mol Brain Res 29, 211-23 (1995).
- Komiyama, N. H. et al. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci 22, 9721-32 (2002).
- Ibrahim, H. M. et al. Ionotropic glutamate receptor binding and subunit mRNA expression in thalamic nuclei in schizophrenia. Am J Psychiatry 157, 1811-23 (2000).
- Whitham, R. H. et al. Lymphocytes from SJL/J mice immunized with spinal cord respond selectively to a peptide of proteolipid protein and transfer relapsing demyelinating experimental autoimmune encephalomyelitis. J Immunol 146, 101-7 (1991).
- Bongarzone, E. R. et al. Identification of a new exon in the myelin proteolipid protein gene encoding novel protein isoforms that are restricted to the somata of oligodendrocytes and neurons. J Neurosci 19, 8349-57 (1999).
- Bruno, R. et al. Multiple sclerosis candidate autoantigens except myelin oligodendrocyte glycoprotein are transcribed in human thymus. Eur J Immunol 32, 2737-47 (2002).
- Jewtoukoff, V., Amzazi, S., Lebar, R., Bach, M. A. & Marche, P. N. T-cell receptor identification of an oligodendrocyte-specific autoreactive cytotoxic T-cell clone without self restriction. Scand J Immunol 36, 893-8 (1992).
- Wakabayashi, K. et al. Phenotypic down-regulation of glutamate receptor subunit GluR1 in Alzheimer's disease. Neurobiol Aging 20, 287-95 (1999).
- Pellegrini-Giampietro, D. E., Bennett, M. V. & Zukin, R. S. AMPA/kainate receptor gene expression in normal and Alzheimer's disease hippocampus. Neuroscience 61, 41-9 (1994).
- Yasuda, R. P. et al. Reduction of AMPA-selective glutamate receptor subunits in the entorhinal cortex of patients with Alzheimer's disease pathology: a biochemical study. Brain Res 678, 161-7 (1995).
- Aronica, E., Dickson, D. W., Kress, Y., Morrison, J. H. & Zukin, R. S. Non-plaque dystrophic dendrites in Alzheimer hippocampus: a new pathological structure revealed by glutamate receptor immunocytochemistry. Neuroscience 82, 979-91 (1998).
- Eder, P., Reinprecht, I., Schreiner, E., Skofitsch, G. & Windisch, M. Increased density of glutamate receptor subunit 1 due to Cerebrolysin treatment: an immunohistochemical study on aged rats. Histochem J 33, 605-12 (2001).
- Gorman, M. P. et al. Steroid-responsive neurologic relapses in a child with a proteolipid protein-1 mutation. Neurology 68, 1305-7 (2007).
- Grigorenko, E. et al. Changes in glutamate receptor subunit composition in hippocampus and cortex in patients with refractory epilepsy. J Neurol Sci 153, 35-45 (1997).
- Babb, T. L. et al. Glutamate AMPA receptors in the fascia dentata of human and kainate rat hippocampal epilepsy. Epilepsy Res 26, 193-205 (1996).
- Gerber, D. J. et al. Evidence for association of schizophrenia with genetic variation in the 8p21.3 gene, PPP3CC, encoding the calcineurin gamma subunit. Proc Natl Acad Sci U S A 100, 8993-8 (2003).
- Beveridge, N. J. et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet 17, 1156-68 (2008).
- Cumming, R. C., Dargusch, R., Fischer, W. H. & Schubert, D. Increase in expression levels and resistance to sulfhydryl oxidation of peroxiredoxin isoforms in amyloid beta-resistant nerve cells. J Biol Chem 282, 30523-34 (2007).
- Ganor, Y. et al. Antibodies to glutamate receptor subtype 3 (GluR3) are found in some patients suffering from epilepsy as the main disease, but not in patients whose epilepsy accompanies antiphospholipid syndrome or Sneddon's syndrome. Autoimmunity 38, 417-24 (2005).
- Watson, R. et al. Absence of antibodies to glutamate receptor type 3 (GluR3) in Rasmussen encephalitis. Neurology 63, 43-50 (2004).
- Levite, M., Fleidervish, I. A., Schwarz, A., Pelled, D. & Futerman, A. H. Autoantibodies to the glutamate receptor kill neurons via activation of the receptor ion channel. J Autoimmun 13, 61-72 (1999).
- Levite, M. & Hermelin, A. Autoimmunity to the glutamate receptor in mice--a model for Rasmussen's encephalitis? J Autoimmun 13, 73-82 (1999).
- Moga, D. E. et al. Glutamate receptor subunit 3 (GluR3) immunoreactivity delineates a subpopulation of parvalbumin-containing interneurons in the rat hippocampus. J Comp Neurol 462, 15-28 (2003).
- Rembach, A. et al. Antisense peptide nucleic acid targeting GluR3 delays disease onset and progression in the SOD1 G93A mouse model of familial ALS. J Neurosci Res 77, 573-82 (2004).
- Fang, J., Nakamura, T., Cho, D. H., Gu, Z. & Lipton, S. A. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson's disease. Proc Natl Acad Sci U S A 104, 18742-7 (2007).
- Wu, Y. et al. Mutations in ionotropic AMPA receptor 3 alter channel properties and are associated with moderate cognitive impairment in humans. Proc Natl Acad Sci U S A 104, 18163-8 (2007).
- Julien, J. P. Amyotrophic lateral sclerosis. unfolding the toxicity of the misfolded. Cell 104, 581-91 (2001).
- Motazacker, M. M. et al. A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am J Hum Genet 81, 7928 (2007).
- Kaukonen, J. et al. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science 289, 782-5 (2000).
- Behan, A., Byrne, C., Dunn, M. J., Cagney, G. & Cotter, D. R. Proteomic analysis of membrane microdomain-associated proteins in the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder reveals alterations in LAMP, STXBP1 and BASP1 protein expression. Mol Psychiatry (2008).
- Bickel, S., Lipp, H. P. & Umbricht, D. Impaired attentional modulation of auditory evoked potentials in N-methyl-D-aspartate NR1 hypomorphic mice. Genes Brain Behav 6, 558-68 (2007).
- Potkin, S. G. et al. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr Bull 35, 96-108 (2009).
- Mundo, E. et al. Evidence that the N-methyl-D-aspartate subunit 1 receptor gene (GRIN1) confers susceptibility to bipolar disorder. Mol Psychiatry 8, 241-5 (2003).
- Begni, S. et al. Association between the G1001C polymorphism in the GRIN1 gene promoter region and schizophrenia. Biol Psychiatry 53, 617-9 (2003).
- Suh, J. G., Ryoo, Z. W., Won, M. H., Oh, Y. S. & Kang, T. C. Differential alteration of NMDA receptor subunits in the gerbil dentate gyrus and subiculum following seizure. Brain Res 904, 104-11 (2001).
- Smith, R. E., Haroutunian, V., Davis, K. L. & Meador-Woodruff, J. H. Expression of excitatory amino acid transporter transcripts in the thalamus of subjects with schizophrenia. Am J Psychiatry 158, 1393-9 (2001).
- Arning, L. et al. NR2A and NR2B receptor gene variations modify age at onset in Huntington disease in a sex-specific manner. Hum Genet 122, 175-82 (2007).
- Cardoso, C. et al. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am J Hum Genet 72, 918-30 (2003).
- Martucci, L. et al. N-methyl-D-aspartate receptor NR2B subunit gene GRIN2B in schizophrenia and bipolar disorder: Polymorphisms and mRNA levels. Schizophr Res 84, 214-21 (2006).
- Abdolmaleky, H. M., Thiagalingam, S. & Wilcox, M. Genetics and epigenetics in major psychiatric disorders: dilemmas, achievements, applications, and future scope. Am J Pharmacogenomics 5, 149-60 (2005).
- Fallin, M. D. et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet 77, 918-36 (2005).
- Knuesel, M. et al. Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol Cell Proteomics 2, 1225-33 (2003).
- Arzberger, T., Krampfl, K., Leimgruber, S. & Weindl, A. Changes of NMDA receptor subunit (NR1, NR2B) and glutamate transporter (GLT1) mRNA expression in Huntington's disease--an in situ hybridization study. J Neuropathol Exp Neurol 56, 440-54 (1997).
Supplementary tables.
Supplementary Table 1. Proteins identified in the single step and tandem purifications by LC-MS/MS.
Approved gene symbols, MGI IDs, UniProt accession numbers, Gene accession numbers in the Genes to Cognition database (G2Cdb) (http://www.genes2cognition.org) and number of approved peptides for each protein identified by LC-MS/MS analysis in the single step and 4 tandem replicates are shown. See Material and Methods for peptide approval criteria. Genes that are also in the MASC/NRC, AMPA receptor (AMPA), metabotrobic glutamate receptor 5 (mGluR5), PSD and PSP lists (Collins et al., 2006) are shown. Common proteins with the PSD-95 immunoprecipitation reported by Dosemeci et al. are also shown (Dosemeci et al., 2007). Using Panther categories (http://www.pantherdb.org), the molecular function and biological process of each protein is indicated. Numbers of peptides from proteins identified in the PSD-95TAP/TAP mice are separated by the symbol | from number of peptides that are also in the wild type purification.
| Number of peptides | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MGI Symbol | Approved Name | MGI ID | UniProt Acc | G2Cdb (mouse) | PSD-95 Core Complex | MASC/NRC | AMPA | mGluR5 | PSD | PSP | Dosemeci et al. | Interactor | Single | T1 | T2 | T3 | T4 | Molecular Function | Biological Process |
| Ablim1 | actin-binding LIM protein 1 | MGI:1194500 | Q8K4G5 | G00000574 | X | X | X | X | 1 | 20 | 10 | 18 | 22 | Cytoeskeletal/ Structural/ Cell adhesion | Actin binding cytoskeletal/Structural protein | ||||
| Abr | active BCR-related gene | MGI:107771 | Q6PCY1 | X | 2 | 2 | 6 | G-protein signaling | Guanyl-nucleotide exchange factor | ||||||||||
| Acat1 | acetyl-Coenzyme A acetyltransferase 1 | MGI:87870 | Q8QZT1 | G00000448 | X | X | X | 3 | 2 | 3 | 3 | Enzymes | Acetyltransferase | ||||||
| Aco2 | aconitase 2, mitochondrial | MGI:87880 | Q99KI0 | G00000936 | X | X | X | X | 7|2 | 3 | 3 | 2 | Enzymes/mitoc | Dehydratase;Hydratase | |||||
| Acot7 | acyl-CoA thioesterase 7 | MGI:1917275 | Q91V12 | X | 5 | 3 | 3 | 3 | 4 | Enzymes | Esterase | ||||||||
| Actn1 | actinin, alpha 1 | MGI:2137706 | A1BN54 | G00000542 | X | X | X | 3 | Cytoeskeletal/ Structural/ Cell adhesion | Non-motor actin binding protein | |||||||||
| Actn4 | actinin alpha 4 | MGI:1890773 | P57780 | G00000544 | X | X | X | X | 2 | Cytoeskeletal/ Structural/ Cell adhesion | Non-motor actin binding protein | ||||||||
| Adam22 | a disintegrin and metallopeptidase domain 22 | MGI:1340046 | Q9R1V6 | G00001236 | X | X | X | X | x | 4 | 12 | 16 | 24 | 31 | Cytoeskeletal/ Structural/ Cell adhesion | Metalloprotease | |||
| AI662250 | expressed sequence AI662250 | MGI:2146912 | Q3UKV2 | X | 2 | 2 | 2 | Unclassified | Unclassified | ||||||||||
| Akr1a4 | aldo-keto reductase family 1, member A4 (aldehyde reductase) | MGI:1929955 | Q80XJ7 | G00001242 | X | X | 3 | Enzymes | Reductase | ||||||||||
| Aldh2 | aldehyde dehydrogenase 2, mitochondrial | MGI:99600 | P47738 | 2 | Enzymes/mitoc | Dehydrogenase | |||||||||||||
| Aldh5a1 | aldhehyde dehydrogenase family 5, subfamily A1 | MGI:2441982 | Q5SZW1 | 3 | Enzymes | Dehydrogenase | |||||||||||||
| Aldoa | aldolase A, fructose-bisphosphate | MGI:87994 | Q6NY00 | G00000917 | X | X | X | X | 11 | Enzymes | Aldolase | ||||||||
| Aldoc | aldolase C, fructose-bisphosphate | MGI:101863 | P05063 | G00000369 | X | X | X | X | X | X | 4 | 2 | 4 | Enzymes | Aldolase | ||||
| Anks1 | ankyrin repeat and SAM domain containing 1 | MGI:2446180 | P59672 | X | 4 | 3 | 3 | Adaptor/ Regulatory | Transmembrane receptor regulatory/adaptor | ||||||||||
| Anks1b | ankyrin repeat and sterile alpha motif domain containing 1B | MGI:1924781 | Q8BZM2 | G00001138 | X | X | X | X | 4 | 17 | 5 | 10 | 10 | Adaptor/ Regulatory | Transmembrane receptor regulatory/adaptor | ||||
| Anxa5 | annexin A5 | MGI:106008 | Q3U8K1 | 2 | Signal transduction | Transfer/carrier protein;Annexin | |||||||||||||
| Ap2a1 | adaptor protein complex AP-2, alpha 1 subunit | MGI:101921 | P17426 | G00000663 | X | X | 3 | Adaptor/ Regulatory | Transmembrane receptor regulatory/adaptor | ||||||||||
| Apoe | apolipoprotein E | MGI:88057 | Q3TXU4 | G00001024 | 2 | Signal transduction | Transporter;Apolipoprotein | ||||||||||||
| Arc | activity regulated cytoskeletal-associated protein | MGI:88067 | Q9WV31 | G00000003 | X | X | X | X | X | 8 | 8 | 13 | 20 | 19 | Cytoeskeletal/ Structural/ Cell adhesion | Unclassified | |||
| Arf1 | ADP-ribosylation factor 1 | MGI:99431 | P84078 | 4 | G-protein signaling | Small GTPase | |||||||||||||
| Arf3 | ADP-ribosylation factor 3 | MGI:99432 | P61205 | G00000363 | X | X | X | 2 | 2 | 3 | 4 | Vesicular/ Trafficking/ Transport | Small GTPase | ||||||
| Arpc4 | actin related protein 2/3 complex, subunit 4 | MGI:1915339 | P59999 | G00000538 | X | X | X | X | 2 | 2 | 4 | Cytoeskeletal/ Structural/ Cell adhesion | Other actin family cytoskeletal protein | ||||||
| Asrgl1 | asparaginase like 1 | MGI:1913764 | Q9CVX3 | 5 | Enzymes | Hydrolase;Other proteases | |||||||||||||
| Atp1b1 | ATPase, Na+/K+ transporting, beta 1 polypeptide | MGI:88108 | P14094 | G00000077 | X | X | X | X | 10 | 3 | 3 | 6 | 3 | Receptors/ Channels/ Transporters | Cation transporter | ||||
| Atp1b2 | ATPase, Na+/K+ transporting, beta 2 polypeptide | MGI:88109 | P14231 | 4 | Receptors/ Channels/ Transporters | Cation transporter | |||||||||||||
| Atp2b1 | ATPase, Ca++ transporting, plasma membrane 1 | MGI:104653 | Q05CJ5 | G00000071 | X | X | X | 8 | 2 | 4 | Receptors/ Channels/ Transporters | Ion channel;Cation transporter;Other hydrolase | |||||||
| Atp2b2 | ATPase, Ca++ transporting, plasma membrane 2 | MGI:105368 | Q3UHH0 | G00000072 | X | X | x | 6 | Receptors/ Channels/ Transporters | Ion channel;Cation transporter;Other hydrolase | |||||||||
| Atp2b4 | ATPase, Ca++ transporting, plasma membrane 4 | MGI:88111 | Q6Q477 | G00000073 | X | X | X | x | 5 | Receptors/ Channels/ Transporters | Cation transport;Calcium ion homeostasis | ||||||||
| Atp4a | ATPase, H+/K+ exchanging, gastric, alpha polypeptide | MGI:88113 | Q91WH7 | 4 | Receptors/ Channels/ Transporters | Ion channel;Cation transporter;Other hydrolase | |||||||||||||
| Atp5a1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 1 | MGI:88115 | Q03265 | G00000299 | X | X | X | X | 5 | 10 | 14 | 11 | Mitochondrial | Unclassified | |||||
| Atp5b | ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit | MGI:107801 | P56480 | G00000296 | X | X | X | 2 | 6 | 5 | Mitochondrial | Ion channel;Hydrogen transporter;ATP synthase | |||||||
| Atp5c1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | MGI:1261437 | Q91VR2 | G00000303 | X | X | X | X | 3 | 3 | 3 | 4 | Receptors/ Channels/ Transporters | Hydrogen transporter;Synthase;Other hydrolase | |||||
| Atp5d | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | MGI:1913293 | Q9DCZ0 | 2 | Receptors/ Channels/ Transporters | Hydrogen transporter;Synthase;Other hydrolase | |||||||||||||
| Atp5o | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | MGI:106341 | Q9DB20 | X | 2 | 2 | 4 | 4 | Mitochondrial | Hydrogen transporter;Synthase;Other hydrolase | |||||||||
| Atp6v0d1 | ATPase, H+ transporting, lysosomal V0 subunit D1 | MGI:1201778 | P51863 | G00000300 | X | X | X | X | X | 4 | 3 | 3 | 4 | 2 | Receptors/ Channels/ Transporters | Hydrogen transporter;Synthase;Other hydrolase | |||
| Atp6v1a | ATPase, H+ transporting, lysosomal V1 subunit A | MGI:1201780 | P50516 | G00000297 | X | X | X | 13 | Receptors/ Channels/ Transporters | Ion channel;Hydrogen transporter;ATP synthase | |||||||||
| Atp6v1b2 | ATPase, H+ transporting, lysosomal V1 subunit B2 | MGI:109618 | Q3TG74 | G00000298 | X | X | X | 5 | Receptors/ Channels/ Transporters | Ion channel;Hydrogen transporter;ATP synthase | |||||||||
| B630019K06Rik | RIKEN cDNA B630019K06 gene | MGI:2147918 | Q7TNS5 | X | 3 | 6 | 7 | 8 | 9 | Unclassified | Unclassified | ||||||||
| Baiap2 | brain-specific angiogenesis inhibitor 1-associated protein 2 | MGI:2137336 | Q8BKX1 | G00000012 | X | X | X | X | x | 14 | 20 | 12 | 13 | 13 | Receptors/ Channels/ Transporters | Receptor | |||
| Basp1 | brain abundant, membrane attached signal protein 1 | MGI:1917600 | Q91XV3 | G00000904 | X | X | 2 | Cytoeskeletal/ Structural/ Cell adhesion | Molecular function unclassified | ||||||||||
| Begain | brain-enriched guanylate kinase-associated | MGI:3044626 | Q68EF6 | G00000870 | X | X | X | X | x | 4 | 8 | 12 | 16 | Adaptor/ Regulatory | Unclassified | ||||
| Btbd11 | BTB (POZ) domain containing 11 | MGI:1921257 | Q6GQW0 | X | 8 | 5 | 3 | 2 | 3 | Signal transduction | Growth factor | ||||||||
| Cacna2d1 | calcium channel, voltage-dependent, alpha2/delta subunit 1 | MGI:88295 | O08532 | G00000856 | X | X | 2 | Receptors/ Channels/ Transporters | Voltage-gated calcium channel | ||||||||||
| Cacng2 | calcium channel, voltage-dependent, gamma subunit 2 | MGI:1316660 | O88602 | G00000911 | X | X | X | X | X | x | 2 | 2 | 2 | 3 | Receptors/ Channels/ Transporters | Voltage-gated calcium channel | |||
| Camk2a | calcium/calmodulin-dependent protein kinase II alpha | MGI:88256 | P11798 | G00000016 | X | X | X | X | X | x | 7|2 | 11 | 3 | 4 | 11 | Ser/Thr Kinase | Non-receptor serine/threonine protein kinase | ||
| Camk2b | calcium/calmodulin-dependent protein kinase II, beta | MGI:88257 | Q5SVI3 | G00000151 | X | X | X | X | X | 7 | 10 | 5 | 5 | 7 | Ser/Thr Kinase | Non-receptor serine/threonine protein kinase | |||
| Camk2d | calcium/calmodulin-dependent protein kinase II, delta | MGI:1341265 | Q6PHZ2 | G00000152 | X | X | X | 4 | Ser/Thr Kinase | Non-receptor serine/threonine protein kinase | |||||||||
| Cap1 | CAP, adenylate cyclase-associated protein 1 (yeast) | MGI:88262 | Q8BPT7 | 2 | Cytoeskeletal/ Structural/ Cell adhesion | Actin binding cytoskeletal protein | |||||||||||||
| Capza2 | capping protein (actin filament) muscle Z-line, alpha 2 | MGI:106222 | P47754 | G00000570 | X | X | X | X | X | 3 | 2 | 2 | 2 | Cytoeskeletal/ Structural/ Cell adhesion | Non-motor actin binding protein | ||||
| Cfl1 | cofilin 1, non-muscle | MGI:101757 | P18760 | G00001165 | X | X | X | 2 | 2 | 3 | 4 | Cytoeskeletal/ Structural/ Cell adhesion | Non-motor actin binding protein | ||||||
| Cit | citron | MGI:105313 | P49025 | G00000240 | X | X | X | X | X | x | 11 | Enzymes | Non-receptor serine/threonine protein kinase | ||||||
| Ckmt1 | creatine kinase, mitochondrial 1, ubiquitous | MGI:99441 | P30275 | G00000440 | X | X | 13 | Kinase | Other kinase | ||||||||||
| Cltc | clathrin, heavy polypeptide (Hc) | MGI:2388633 | Q5SXR6 | G00000656 | X | X | X | X | X | 24 | 10 | 4 | 12 | Vesicular/ Trafficking/ Transport | Vesicle coat protein | ||||
| Clu | clusterin | MGI:88423 | Q06890 | G00000645 | X | X | 2 | 3 | Unclassified | Unclassified | |||||||||
| Cnp | 2',3'-cyclic nucleotide 3' phosphodiesterase | MGI:88437 | P16330 | G00000901 | X | X | X | 11 | 10 | 3 | 11 | 10 | Enzymes | Phosphodiesterase | |||||
| Cntn1 | contactin 1 | MGI:105980 | P12960 | G00000628 | X | X | 2 | Cytoeskeletal/ Structural/ Cell adhesion | CAM family adhesion molecule | ||||||||||
| Cpne4* | copine IV | MGI:1921270 | Q8BLR2 | X | 4 | 2 | 3 | Vesicular/ Trafficking/ Transport | Miscellaneus function protein;Membrane traffic | ||||||||||
| Cpne7 | copine VII | MGI:2142747 | Q0VE82 | X | 4 | 3 | 2 | 3 | Vesicular/ Trafficking/ Transport | Miscellaneus function protein;Membrane traffic | |||||||||
| Crym | crystallin, mu | MGI:102675 | O54983 | 2 | 3 | 3 | Enzymes | Other lyase | |||||||||||
| Cs | citrate synthase | MGI:88529 | Q9CZU6 | G00000940 | X | X | 8 | Enzymes | Synthase;Transferase;Other lyase | ||||||||||
| Csl | citrate synthase like | MGI:1919082 | Q9DAM4 | 4 | Unclassified | Molecular function unclassified | |||||||||||||
| Csmd2 | CUB and Sushi multiple domains 2 | MGI:2386401 | A2A8D7 | 5 | Enzymes | Acyltransferase | |||||||||||||
| Cycs | cytochrome c, somatic | MGI:88578 | P62897 | 3 | Mitochondrial | Mitochondrial carrier protein | |||||||||||||
| Cyfip2 | cytoplasmic FMR1 interacting protein 2 | MGI:1924134 | Q924D3 | G00000470 | X | X | X | 2 | G-protein signaling | Other G-protein modulator | |||||||||
| Dbi | diazepam binding inhibitor | MGI:94865 | Q548W7 | 2 | Receptors/ Channels/ Transporters | Other receptor;Other transporter | |||||||||||||
| Ddah1 | dimethylarginine dimethylaminohydrolase 1 | MGI:1916469 | Q3UF65 | 2 | Enzymes | Other hydrolase | |||||||||||||
| Dld | dihydrolipoamide dehydrogenase | MGI:107450 | O08749 | G00000459 | X | X | 3 | Enzymes | Dehydrogenase | ||||||||||
| Dlg1 | discs, large homolog 1 (Drosophila) | MGI:107231 | Q3UP61 | G00000863 | X | X | X | X | X | 17 | 24 | 42 | 34 | 42 | Adaptor/ Regulatory | Transmembrane receptor regulatory/adaptor | |||
| Dlg2 | discs, large homolog 2 (Drosophila) | MGI:1344351 | Q91XM9 | G00000010 | X | X | X | X | X | x | 42 | 49 | 67 | 69 | 80 | Adaptor/ Regulatory | Unclassified | ||
| Dlg3 | discs, large homolog 3 (Drosophila) | MGI:1888986 | Q52KF7 | G00000007 | X | X | X | X | X | x | 16 | 27 | 17 | 14 | 22 | Adaptor/ Regulatory | Transmembrane receptor regulatory/adaptor | ||
| Dlg4 | discs, large homolog 4 (Drosophila) | MGI:1277959 | Q62108 | G00000004 | X | X | X | X | X | x | 45 | 42 | 57 | 64 | 64 | Adaptor/ Regulatory | Transmembrane receptor regulatory/adaptor | ||
| Dlgap1 | discs, large (Drosophila) homolog-associated protein 1 | MGI:1346065 | Q9D415 | G00000869 | X | X | X | X | X | x | 10 | 11 | 9 | 14 | 24 | Adaptor/ Regulatory | Transmembrane receptor regulatory/adaptor | ||
| Dlgap2 | discs, large (Drosophila) homolog-associated protein 2 | MGI:2443181 | Q8BJ42 | G00000132 | X | X | X | X | x | 9 | 18 | 12 | 19 | 24 | Adaptor/ Regulatory | Transmembrane receptor regulatory/adaptor | |||
| Dlgap3 | discs, large (Drosophila) homolog-associated protein 3 | MGI:3039563 | A2A7T7 | X | x | 3 | 16 | 8 | 7 | 15 | Adaptor/ Regulatory | Transmembrane receptor regulatory/adaptor | |||||||
| Dlgap4 | discs, large homolog-associated protein 4 (Drosophila) | MGI:2138865 | A2BDU3 | G00000872 | X | X | X | X | x | 12 | 14 | 9 | 7 | 14 | Adaptor/ Regulatory | Transmembrane receptor regulatory/adaptor | |||
| Dstn | destrin | MGI:1929270 | Q9R0P5 | X | 3 | 2 | 5 | 6 | 6 | Cytoeskeletal/ Structural/ Cell adhesion | Non-motor actin binding protein | ||||||||
| Dusp10 | dual specificity phosphatase 10 | MGI:1927070 | Q8R3L3 | G00000202 | X | X | 3 | 2 | Phosphatase | Kinase inhibitor;Protein phosphatase | |||||||||
| Dync1h1 | dynein cytoplasmic 1 heavy chain 1 | MGI:103147 | Q9JHU4 | G00000758 | X | X | 3 | Vesicular/ Trafficking/ Transport | Microtubule binding motor protein;Hydrolase | ||||||||||
| Dynll1 | dynein light chain LC8-type 1 | MGI:1861457 | Q3UGE7 | G00000759 | X | X | X | x | 5 | Cytoeskeletal/ Structural/ Cell adhesion | Cytoskeletal protein | ||||||||
| Eef1a1* | eukaryotic translation elongation factor 1 alpha 1 | MGI:1096881 | P10126 | G00000505 | X | X | X | 5 | 2 | 2 | Transcription/ Translation | Translation elongation factor | |||||||
| Eef1g | eukaryotic translation elongation factor 1 gamma | MGI:1914410 | Q9D8N0 | G00000507 | X | X | 4 | Transcription/ Translation | Translation elongation factor;Cytoskeletal | ||||||||||
| Eif4a1 | eukaryotic translation initiation factor 4A1 | MGI:95303 | Q3TFG3 | 4 | Transcription/ Translation | RNA helicase;Translation initiation factor | |||||||||||||
| Eif4a2 | eukaryotic translation initiation factor 4A2 | MGI:106906 | P10630 | G00000956 | X | X | 5 | Transcription/ Translation | RNA helicase;Translation initiation factor | ||||||||||
| Eno1 | enolase 1, alpha non-neuron | MGI:95393 | Q5XJG8 | G00001203 | X | X | X | 15 | Enzymes | Lyase | |||||||||
| Eno3 | enolase 3, beta muscle | MGI:95395 | Q4FK59 | G00000403 | X | X | X | 3 | Enzymes | Lyase | |||||||||
| Fabp3 | fatty acid binding protein 3, muscle and heart | MGI:95476 | Q5EBJ0 | 2 | Receptors/ Channels/ Transporters | Other transfer/carrier protein | |||||||||||||
| Fah | fumarylacetoacetate hydrolase | MGI:95482 | Q3TY87 | 2 | Enzymes | Other hydrolase | |||||||||||||
| Fam81a | family with sequence similarity 81, member A | MGI:1924136 | Q3UXZ6 | G00000816 | X | X | X | X | 5 | 7 | 6 | 21 | 18 | Unclassified | Unclassified | ||||
| Fbxo2 | F-box protein 2 | MGI:2446216 | Q80UW2 | G00000489 | X | X | 4 | Signal transduction | Other miscellaneous function protein | ||||||||||
| Fbxo6 | F-box protein 6 | MGI:1354743 | Q9QZN4 | 3 | 4 | Signal transduction | Ubiquitin-protein ligase | ||||||||||||
| Fh1 | fumarate hydratase 1 | MGI:95530 | Q3UIA9 | 2 | Enzymes | Other lyase | |||||||||||||
| Fkbp1a | FK506 binding protein 1a | MGI:95541 | Q3ULN5 | 3 | Enzymes | Other isomerase | |||||||||||||
| Frmpd3 | FERM and PDZ domain containing 3 | MGI:3646547 | Q8BXG0 | X | 2 | 5 | 5 | Unclassified | Unclassified | ||||||||||
| Fscn1 | fascin homolog 1, actin bundling protein (Strongylocentrotus purpuratus) | MGI:1352745 | Q61553 | G00000612 | X | X | X | 4 | 2 | 2 | 3 | Cytoeskeletal/ Structural/ Cell adhesion | Non-motor actin binding protein | ||||||
| Gapdh | glyceraldehyde-3-phosphate dehydrogenase | MGI:95640 | P16858 | G00000935 | X | X | X | X | X | 8 | 9 | 8 | 9 | 10 | Enzymes | Unclassified | |||
| Gda | guanine deaminase | MGI:95678 | Q9R111 | G00000015 | X | x | 29|4 | 29 | 26 | 29 | 31 | Enzymes | Other hydrolase | ||||||
| Gdi1 | guanosine diphosphate (GDP) dissociation inhibitor 1 | MGI:99846 | P50396 | G00000254 | X | X | X | 5 | G-protein signaling | G-protein modulator;Acyltransferase | |||||||||
| Glod4 | glyoxalase domain containing 4 | MGI:1914451 | Q9CY26 | 2 | Enzymes | Other lyase | |||||||||||||
| Glud1 | glutamate dehydrogenase 1 | MGI:95753 | Q8C273 | G00000442 | X | X | 5 | Enzymes | Dehydrogenase | ||||||||||
| Glul | glutamate-ammonia ligase (glutamine synthetase) | MGI:95739 | P15105 | G00000918 | X | X | X | X | X | 8 | 5 | 17 | 21 | 19 | Enzymes | Synthetase;Other ligase | |||
| Gna13 | guanine nucleotide binding protein, alpha 13 | MGI:95768 | Q8C5L2 | G00001111 | 2 | 2 | G-protein signalling | Large G-protein | |||||||||||
| Gnai1 | guanine nucleotide binding protein (G protein), alpha inhibiting 1 | MGI:95771 | B2RSH2 | G00001151 | 4 | G-protein signaling | Large G-protein | ||||||||||||
| Gnai2 | guanine nucleotide binding protein (G protein), alpha inhibiting 2 | MGI:95772 | P08752 | G00000215 | X | X | 5 | G-protein signaling | Large G-protein | ||||||||||
| Gnao1 | guanine nucleotide binding protein, alpha O | MGI:95775 | P18872 | G00000208 | X | X | X | X | X | 10 | 8 | 7 | 15 | 16 | G-protein signaling | Large G-protein | |||
| Gnaq | guanine nucleotide binding protein, alpha q polypeptide | MGI:95776 | Q3UHH5 | G00001143 | X | X | 2 | G-protein signaling | Large G-protein | ||||||||||
| Gnat2 | guanine nucleotide binding protein, alpha transducing 2 | MGI:95779 | P50149 | G00001226 | 3 | G-protein signaling | Large G-protein | ||||||||||||
| Gnb1 | guanine nucleotide binding protein (G protein), beta 1 | MGI:95781 | P62874 | G00000210 | X | X | X | 3 | 3 | 3 | G-protein signaling | Large G-protein;Hydrolase | |||||||
| Gnb4 | guanine nucleotide binding protein (G protein), beta 4 | MGI:104581 | Q3TJJ1 | G00000235 | X | X | 2 | G-protein signaling | Large G-protein;Hydrolase | ||||||||||
| Got1 | glutamate oxaloacetate transaminase 1, soluble | MGI:95791 | Q3UJH8 | 13 | Enzymes | Molecular function unclassified | |||||||||||||
| Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | MGI:95679 | P13707 | 2 | Enzymes | Dehydrogenase | |||||||||||||
| Gpd2 | glycerol phosphate dehydrogenase 2, mitochondrial | MGI:99778 | Q64521 | G00000942 | X | X | 7 | Enzymes/mitoc | Dehydrogenase | ||||||||||
| Gpi1 | glucose phosphate isomerase 1 | MGI:95797 | Q3UUX1 | 9 | Enzymes | Glycosyltransferase | |||||||||||||
| Gpm6a | glycoprotein m6a | MGI:107671 | Q8R1P3 | G00000560 | X | X | X | 2 | Cytoeskeletal/ Structural/ Cell adhesion | Myelin protein | |||||||||
| Gpr123 | G protein-coupled receptor 123 | MGI:1277167 | Q52KJ6 | X | 2 | 3 | 3 | 3 | Receptors/ Channels/ Transporters | G-protein coupled receptor | |||||||||
| Gpx4 | glutathione peroxidase 4 | MGI:104767 | O70325 | G00000943 | X | X | X | 2 | 2 | 2 | Enzymes | Peroxidase | |||||||
| Grb2 | growth factor receptor bound protein 2 | MGI:95805 | Q60631 | G00000139 | X | X | X | 2 | 4 | Adaptor/ Regulatory | Signaling;miscellaneous;regulatory/adaptor | ||||||||
| Gria1 | glutamate receptor, ionotropic, AMPA1 (alpha 1) | MGI:95808 | Q5NBY1 | G00000013 | X | X | X | X | X | 2 | 3 | 5 | 13 | Receptors/ Channels/ Transporters | Glutamate receptor;Ion channel | ||||
| Gria2 | glutamate receptor, ionotropic, AMPA2 (alpha 2) | MGI:95809 | P23819 | G00000058 | X | X | X | X | X | 4 | 4 | 8 | 10 | 20 | Receptors/ Channels/ Transporters | Glutamate receptor;Ion channel | |||
| Gria3 | glutamate receptor, ionotropic, AMPA3 (alpha 3) | MGI:95810 | Q9Z2W9 | G00000059 | X | X | X | X | X | 3 | 2 | 9 | 18 | Receptors/ Channels/ Transporters | Glutamate receptor;Ion channel | ||||
| Gria4 | glutamate receptor, ionotropic, AMPA4 (alpha 4) | MGI:95811 | Q9Z2W8 | G00000060 | X | X | X | X | X | 2 | 2 | 5 | 9 | Receptors/ Channels/ Transporters | Glutamate receptor;Ion channel | ||||
| Grik2 | glutamate receptor, ionotropic, kainate 2 (beta 2) | MGI:95815 | P39087 | G00000063 | X | X | X | X | X | x | 2 | 4 | 6 | Receptors/ Channels/ Transporters | Glutamate receptor;Ion channel | ||||
| Grik5 | glutamate receptor, ionotropic, kainate 5 (gamma 2) | MGI:95818 | Q61626 | G00001115 | X | x | 2 | 3 | 7 | Receptors/ Channels/ Transporters | Glutamate receptor;Ion channel | ||||||||
| Grin1 | glutamate receptor, ionotropic, NMDA1 (zeta 1) | MGI:95819 | A2AI21 | G00000849 | X | X | X | X | X | x | 31 | 29 | 40 | 50 | 55 | Receptors/ Channels/ Transporters | Glutamate receptor;Ion channel | ||
| Grin2a | glutamate receptor, ionotropic, NMDA2A (epsilon 1) | MGI:95820 | P35436 | G00000001 | X | X | X | X | X | X | x | 15 | 24 | 31 | 36 | 46 | Receptors/ Channels/ Transporters | Glutamate receptor;Ion channel | |
| Grin2b | glutamate receptor, ionotropic, NMDA2B (epsilon 2) | MGI:95821 | Q01097 | G00000002 | X | X | X | X | X | x | 32 | 44 | 54 | 67 | 78 | Receptors/ Channels/ Transporters | Glutamate receptor;Ion channel | ||
| Grin2c | glutamate receptor, ionotropic, NMDA2C (epsilon 3) | MGI:95822 | Q01098 | G00001133 | x | 5 | Receptors/ Channels/ Transporters | Glutamate receptor;Ion channel | |||||||||||
| Grin2d | glutamate receptor, ionotropic, NMDA2D (epsilon 4) | MGI:95823 | Q03391 | G00000066 | X | X | X | x | 2 | 3 | 6 | 9 | 10 | Receptors/ Channels/ Transporters | Glutamate receptor;Ion channel | ||||
| Grm3 | glutamate receptor, metabotropic 3 | MGI:1351340 | Q9QYS2 | G00000062 | X | X | 2 | Receptors/ Channels/ Transporters | G-protein coupled receptor | ||||||||||
| Gstp1 | glutathione S-transferase, pi 1 | MGI:95865 | P19157 | 6 | Enzymes | Other transferase | |||||||||||||
| Hist1h2bb* | histone 1, H2bb | MGI:2448377 | Q8CGP2 | 3 | 3 | DNA binding | Unclassified | ||||||||||||
| Hpcal1 | hippocalcin-like 1 | MGI:1855689 | P62748 | 4 | Signal transduction | Calmodulin related protein | |||||||||||||
| Hrsp12 | heat-responsive protein 12 | MGI:1095401 | Q569N4 | 2 | Unclassified | Other miscellaneous function protein | |||||||||||||
| Hsp90aa1 | heat shock protein 90, alpha (cytosolic), class A member 1 | MGI:96250 | Q3UIF3 | 7 | Chaperone/ Protein folding/ Signalling | Hsp 90 family chaperone | |||||||||||||
| Hsp90ab1 | heat shock protein 90 alpha (cytosolic), class B member 1 | MGI:96247 | Q3UIQ7 | G00000269 | X | X | X | 12 | Chaperone/ Protein folding/ Signalling | Hsp 90 family chaperone | |||||||||
| Hspa12a | heat shock protein 12A | MGI:1920692 | Q8K0U4 | G00000276 | X | X | 3 | 2 | Chaperone/ Protein folding/ Signalling | Hsp 70 family chaperone | |||||||||
| Hspa1l | heat shock protein 1-like | MGI:96231 | A1L347 | 3 | Chaperone/ Protein folding/ Signalling | Hsp 70 family chaperone | |||||||||||||
| Hspa2 | heat shock protein 2 | MGI:96243 | Q99KD7 | 8 | Chaperone/ Protein folding/ Signalling | Hsp 70 family chaperone | |||||||||||||
| Hspa5 | heat shock protein 5 | MGI:95835 | Q3TKF8 | G00000266 | X | X | X | 8 | Chaperone/ Protein folding/ Signalling | Hsp 70 family chaperone | |||||||||
| Hspa8 | heat shock protein 8 | MGI:105384 | Q3U7D7 | X | 24|4 | Chaperone/ Protein folding/ Signalling | Hsp 70 family chaperone | ||||||||||||
| Hspa9 | heat shock protein 9 | MGI:96245 | P38647 | G00000268 | X | X | 12 | Chaperone/ Protein folding/ Signalling | Hsp 70 family chaperone | ||||||||||
| Hspd1 | heat shock protein 1 (chaperonin) | MGI:96242 | Q3UIP0 | G00000275 | X | X | 8 | Chaperone/ Protein folding/ Signalling | Chaperonin | ||||||||||
| Hspe1-rs1 | heat shock protein 1 (chaperonin 10), related sequence 1 | MGI:1935159 | Q9JI95 | 2 | Chaperone/ Protein folding/ Signalling | ||||||||||||||
| Idh3a | isocitrate dehydrogenase 3 (NAD+) alpha | MGI:1915084 | Q9D6R2 | G00000464 | X | X | 5 | Enzymes | Dehydrogenase | ||||||||||
| Immt | inner membrane protein, mitochondrial | MGI:1923864 | Q8CAQ8 | G00000754 | X | X | X | 3 | Unclassified | Molecular function unclassified | |||||||||
| Impa1 | inositol (myo)-1(or 4)-monophosphatase 1 | MGI:1933158 | O55023 | G00000413 | X | X | 3 | Enzymes | Other phosphatase | ||||||||||
| Ina | internexin neuronal intermediate filament protein, alpha | MGI:96568 | Q8VCW5 | G00000566 | X | X | X | X | 4 | Cytoeskeletal/ Structural/ Cell adhesion | Intermediate filament;Structural protein | ||||||||
| Iqsec1 | IQ motif and Sec7 domain 1 | MGI:1196356 | Q8R0S2 | G00001160 | X | X | X | X | 3 | 11 | 7 | 6 | 15 | Vesicular/ Trafficking/ Transport | Guanyl-nucleotide exchange factor | ||||
| Iqsec2 | IQ motif and Sec7 domain 2 | MGI:3528396 | Q5DU25 | G00000373 | X | X | X | X | 10 | 34 | 25 | 24 | 36 | Vesicular/ Trafficking/ Transport | Guanyl-nucleotide exchange factor | ||||
| Jak3 | Janus kinase 3 | MGI:99928 | Q62137 | 2 | Kinase | Non-receptor tyrosine protein kinase | |||||||||||||
| Kalrn | kalirin, RhoGEF kinase | MGI:2685385 | A2CG52 | G00001298 | X | X | X | X | X | 2 | 3 | 2 | 8 | G-protein signaling | Guanyl-nucleotide exchange factor | ||||
| Kcna1 | potassium voltage-gated channel, shaker-related subfamily, member 1 | MGI:96654 | P16388 | X | x | 2 | 6 | 5 | 5 | 6 | Receptors/ Channels/ Transporters | Voltage-gated potassium channel | |||||||
| Kcna2 | potassium voltage-gated channel, shaker-related subfamily, member 2 | MGI:96659 | P63141 | G00000088 | X | X | X | x | 3 | 4 | 5 | 7 | 6 | Receptors/ Channels/ Transporters | Voltage-gated potassium channel | ||||
| Kcna3 | potassium voltage-gated channel, shaker-related subfamily, member 3 | MGI:96660 | P16390 | X | x | 1 | 3 | 4 | 6 | 5 | Receptors/ Channels/ Transporters | Voltage-gated potassium channel | |||||||
| Kcna4 | potassium voltage-gated channel, shaker-related subfamily, member 4 | MGI:96661 | Q8CBF8 | X | x | 2 | 3 | 5 | 5 | Receptors/ Channels/ Transporters | Voltage-gated potassium channel | ||||||||
| Kcnab1 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | MGI:109155 | P63143 | G00001177 | X | 3 | 3 | 4 | 6 | Receptors/ Channels/ Transporters | Voltage-gated potassium channel | ||||||||
| Kcnab2 | potassium voltage-gated channel, shaker-related subfamily, beta member 2 | MGI:109239 | P62482 | G00000857 | X | X | X | 4 | 5 | 6 | 10 | 11 | Receptors/ Channels/ Transporters | Voltage-gated potassium channel | |||||
| Kcnj10 | potassium inwardly-rectifying channel, subfamily J, member 10 | MGI:1194504 | Q9JM63 | X | x | 5 | 3 | 3 | 4 | 6 | Receptors/ Channels/ Transporters | Voltage-gated potassium channel | |||||||
| Kcnj16 | potassium inwardly-rectifying channel, subfamily J, member 16 | MGI:1314842 | Q9Z307 | 3 | 7 | Receptors/ Channels/ Transporters | Voltage-gated potassium channel | ||||||||||||
| Kcnj4 | potassium inwardly-rectifying channel, subfamily J, member 4 | MGI:104743 | P52189 | G00000089 | X | X | X | X | x | 1 | 4 | 6 | 6 | 8 | Receptors/ Channels/ Transporters | Voltage-gated potassium channel | |||
| Lancl1 | LanC (bacterial lantibiotic synthetase component C)-like 1 | MGI:1336997 | O89112 | 3 | 4 | Unclassified | Unclassified | ||||||||||||
| Lancl2 | LanC (bacterial lantibiotic synthetase component C)-like 2 | MGI:1919085 | Q9JJK2 | G00001045 | X | X | 2 | Unclassified | Molecular function unclassified | ||||||||||
| Lap3 | leucine aminopeptidase 3 | MGI:1914238 | Q9CPY7 | 3 | Enzymes | Metalloprotease | |||||||||||||
| Ldha | lactate dehydrogenase A | MGI:96759 | Q3TI99 | G00000415 | X | X | X | 9 | Enzymes | Dehydrogenase | |||||||||
| Ldhb | lactate dehydrogenase B | MGI:96763 | Q545Y4 | G00000416 | X | X | X | 14 | Enzymes | Dehydrogenase | |||||||||
| Lgi1 | leucine-rich repeat LGI family, member 1 | MGI:1861691 | Q9JIA1 | G00000633 | X | X | X | X | 8 | 6 | 12 | 19 | 15 | Receptors/ Channels/ Transporters | Receptor;Extracellular matrix | ||||
| Lrp1 | low density lipoprotein receptor-related protein 1 | MGI:96828 | Q91ZX7 | G00000110 | X | X | X | x | 2 | 3 | Receptors/ Channels/ Transporters | Other receptor | |||||||
| Lrrtm1 | leucine rich repeat transmembrane neuronal 1 | MGI:2389173 | Q8K377 | G00000905 | X | X | X | 2 | Receptors/ Channels/ Transporters | Receptor;Extracellular matrix | |||||||||
| Lsm11 | U7 snRNP-specific Sm-like protein LSM11 | MGI:1919540 | Q8BUV6 | 2 | 3 | Transcription/Translation | mRNA splicing factor | ||||||||||||
| Mapk1 | mitogen-activated protein kinase 1 | MGI:1346858 | P63085 | G00000170 | X | X | X | X | 5 | 3 | 3 | 4 | 9 | Ser/Thr Kinase | Non-receptor serine/threonine protein kinase | ||||
| Mapk3 | mitogen-activated protein kinase 3 | MGI:1346859 | Q63844 | G00000179 | X | X | 4 | Ser/Thr Kinase | Non-receptor serine/threonine protein kinase | ||||||||||
| Mog | myelin oligodendrocyte glycoprotein | MGI:97435 | Q61885 | G00001025 | X | X | X | 2 | 2 | Unclassified | Myelin protein | ||||||||
| Msrb2 | methionine sulfoxide reductase B2 | MGI:1923717 | Q78J03 | X | 2 | 7 | 4 | Enzymes | Reductase | ||||||||||
| Mtap1a | microtubule-associated protein 1 A | MGI:1306776 | Q9QYR6 | G00001073 | X | X | X | 3 | 2 | Cytoeskeletal/ Structural/ Cell adhesion | Non-motor microtubule binding protein | ||||||||
| Mtpn | myotrophin | MGI:99445 | Q543M6 | 2 | Unclassified | Molecular function unclassified | |||||||||||||
| Myl6 | myosin, light polypeptide 6, alkali, smooth muscle and non-muscle | MGI:109318 | Q60605 | G00000768 | X | X | X | 3 | Cytoeskeletal/ Structural/ Cell adhesion | Actin binding protein;Calmodulin related | |||||||||
| Napa | N-ethylmaleimide sensitive fusion protein attachment protein alpha | MGI:104563 | Q543I3 | G00000706 | X | X | 2 | Vesicular/ Trafficking/ Transport | Other membrane traffic protein | ||||||||||
| Napb | N-ethylmaleimide sensitive fusion protein attachment protein beta | MGI:104562 | P28663 | G00000680 | X | X | 3 | Vesicular/ Trafficking/ Transport | Aspartic protease | ||||||||||
| Napg | N-ethylmaleimide sensitive fusion protein attachment protein gamma | MGI:104561 | Q8C1T5 | G00000987 | X | X | 3 | Vesicular/ Trafficking/ Transport | Other membrane traffic protein | ||||||||||
| Ncald | neurocalcin delta | MGI:1196326 | Q91X97 | 2 | Signal transduction | Calmodulin related protein | |||||||||||||
| Ncdn | neurochondrin | MGI:1347351 | Q9Z0E0 | G00001033 | X | X | 4 | Cytoeskeletal/ Structural/ Cell adhesion | Extracellular matrix structural protein | ||||||||||
| Ndufs2 | NADH dehydrogenase (ubiquinone) Fe-S protein 2 | MGI:2385112 | Q91WD5 | G00000313 | X | X | 2 | Enzymes | Dehydrogenase;Reductase | ||||||||||
| Nefl | neurofilament, light polypeptide | MGI:97313 | P08551 | G00000568 | X | X | X | X | X | 7 | 3 | 3 | 6 | Cytoeskeletal/ Structural/ Cell adhesion | Intermediate filament;Structural protein | ||||
| Nefm | neurofilament, medium polypeptide | MGI:97314 | Q3HRJ6 | G00000567 | X | X | X | 2 | Cytoeskeletal/ Structural/ Cell adhesion | Intermediate filament;Structural protein | |||||||||
| Neto1 | neuropilin (NRP) and tolloid (TLL)-like 1 | MGI:2180216 | Q8R4I7 | 2 | 2 | Unclassified | Unclassified | ||||||||||||
| Nlgn2 | neuroligin 2 | MGI:2681835 | Q69ZK9 | G00000635 | X | X | X | x | 2 | Cytoeskeletal/ Structural/ Cell adhesion | Other signaling molecule | ||||||||
| Nlgn3 | neuroligin 3 | MGI:2444609 | A2AGI2 | G00000636 | X | X | X | x | 2 | Cytoeskeletal/ Structural/ Cell adhesion | Other signaling molecule | ||||||||
| Nme1 | non-metastatic cells 1, protein (NM23A) expressed in | MGI:97355 | P15532 | 5 | Kinase | Nucleotide kinase | |||||||||||||
| Npepps | aminopeptidase puromycin sensitive | MGI:1101358 | Q5PR74 | G00000392 | X | X | 2 | Enzymes | Metalloprotease | ||||||||||
| Nptn | neuroplastin | MGI:108077 | P97300 | G00000563 | X | X | 2 | Adaptor/ Regulatory | Transmembrane receptor regulatory/adaptor | ||||||||||
| Nrxn1 | neurexin I | MGI:1096391 | Q9CS84 | G00001103 | X | X | X | X | 8 | 17 | 12 | 23 | Cytoeskeletal/ Structural/ Cell adhesion | Other receptor | |||||
| Nsf | N-ethylmaleimide sensitive fusion protein | MGI:104560 | P46460 | G00000661 | X | X | X | X | X | X | 16|2 | 4 | 4 | 7 | 9 | Enzymes | Other hydrolase | ||
| Nsfl1c | NSFL1 (p97) cofactor (p47) | MGI:3042273 | Q3UVN5 | 4 | Vesicular/ Trafficking/ Transport | Membrane traffic regulatory protein | |||||||||||||
| Nxph3 | neurexophilin 3 | MGI:1336188 | Q91VX5 | 2 | Signal transduction | Signaling molecule | |||||||||||||
| Otub1 | OTU domain, ubiquitin aldehyde binding 1 | MGI:2147616 | Q3V408 | 3 | Enzymes | Other hydrolase | |||||||||||||
| Oxct1 | 3-oxoacid CoA transferase 1 | MGI:1914291 | Q9D0K2 | 2 | Enzymes | Other transferase | |||||||||||||
| Pacsin1 | protein kinase C and casein kinase substrate in neurons 1 | MGI:1345181 | Q61644 | G00000681 | X | X | 2 | Vesicular/ Trafficking/ Transport | Membrane traffic regulatory protein | ||||||||||
| Park7 | Parkinson disease (autosomal recessive, early onset) 7 | MGI:2135637 | A2A817 | X | 4 | 3 | 3 | 3 | Transcription/ Translation | Other RNA-binding protein | |||||||||
| Pcbp1 | poly(rC) binding protein 1 | MGI:1345635 | P60335 | G00000499 | X | X | X | 3 | 3 | 3 | 5 | Transcription/ Translation | Select regulatory molecule | ||||||
| Pcbp2 | poly(rC) binding protein 2 | MGI:108202 | Q61990 | 2 | 3 | 3 | Transcription/ Translation | Select regulatory molecule | |||||||||||
| Pdha1 | pyruvate dehydrogenase E1 alpha 1 | MGI:97532 | P35486 | G00000468 | X | X | X | 3 | 4 | 3 | 8 | 5 | Enzymes | Dehydrogenase | |||||
| Pdhb | pyruvate dehydrogenase (lipoamide) beta | MGI:1915513 | Q9D051 | G00000469 | X | X | X | 5 | 4 | 4 | 4 | 6 | Enzymes | Dehydrogenase | |||||
| Pdia3 | protein disulfide isomerase associated 3 | MGI:95834 | Q99LF6 | 3 | Enzymes | Other isomerase | |||||||||||||
| Pdia6 | protein disulfide isomerase associated 6 | MGI:1919103 | Q3TML0 | G00000286 | X | X | 2 | Enzymes | Other isomerase | ||||||||||
| Pdxk | pyridoxal (pyridoxine, vitamin B6) kinase | MGI:1351869 | Q8K183 | 4 | Kinase | Other kinase | |||||||||||||
| Pebp1 | phosphatidylethanolamine binding protein 1 | MGI:1344408 | Q5EBQ2 | 3 | Signal transduction | Transfer/carrier protein;Kinase inhibitor | |||||||||||||
| Pfkm | phosphofructokinase, muscle | MGI:97548 | Q543L1 | G00000386 | X | X | 4 | Kinase | Carbohydrate kinase | ||||||||||
| Pfkp | phosphofructokinase, platelet | MGI:1891833 | Q8C605 | G00000368 | X | X | 2 | Kinase | Carbohydrate kinase | ||||||||||
| Pfn1 | profilin 1 | MGI:97549 | Q8CEH8 | 4 | Unclassified | Molecular function unclassified | |||||||||||||
| Pgam5 | phosphoglycerate mutase family member 5 | MGI:1919792 | Q3UK19 | G00000810 | X | X | X | 7 | 7 | 8 | 10 | Unclassified | Unclassified | ||||||
| Pgk1 | phosphoglycerate kinase 1 | MGI:97555 | P09411 | X | 11 | 2 | 3 | 13 | 8 | Enzymes | Carbohydrate kinase | ||||||||
| Pgk2 | phosphoglycerate kinase 2 | MGI:97563 | P09041 | 4 | Enzymes | Carbohydrate kinase | |||||||||||||
| Pgm2l1 | phosphoglucomutase 2-like 1 | MGI:1918224 | Q8CAA7 | 3 | Enzymes | Mutase | |||||||||||||
| Phb | prohibitin | MGI:97572 | Q3TV00 | 3 | Unclassified | Molecular function unclassified | |||||||||||||
| Phb2 | prohibitin 2 | MGI:102520 | O35129 | G00001123 | X | X | X | X | 3 | 2 | 5 | 4 | Transcription/ Translation | Unclassified | |||||
| Phyhip | phytanoyl-CoA hydroxylase interacting protein | MGI:1860417 | Q8K0S0 | 2 | Unclassified | Molecular function unclassified | |||||||||||||
| Pkm2 | pyruvate kinase, muscle | MGI:97591 | P52480 | X | 11 | 7 | 6 | 11 | 12 | Enzumes | Carbohydrate kinase | ||||||||
| Plp1 | proteolipid protein (myelin) 1 | MGI:97623 | P60202 | G00000557 | X | X | X | X | X | 2 | 2 | 3 | 4 | 5 | Cytoeskeletal/ Structural/ Cell adhesion | Myelin protein | |||
| Ppap2b | phosphatidic acid phosphatase type 2B | MGI:1915166 | Q99JY8 | G00000428 | X | X | X | 2 | 2 | 5 | Enzymes | Other phosphatase | |||||||
| Ppia | peptidylprolyl isomerase A | MGI:97749 | P52480 | 4 | Enzymes | Other isomerase | |||||||||||||
| Ppp1r7 | protein phosphatase 1, regulatory (inhibitor) subunit 7 | MGI:1913635 | Q62079 | 4 | Phosphatase | Phosphatase modulator | |||||||||||||
| Ppp2r1a | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), alpha isoform | MGI:1926334 | Q7TMX2 | G00000195 | X | X | X | 5 | Phosphatase | Protein phosphatase | |||||||||
| Ppp3ca | protein phosphatase 3, catalytic subunit, alpha isoform | MGI:107164 | P63328 | G00000194 | X | X | X | X | 15|2 | 12 | 6 | 7 | 11 | Phosphatase | Protein phosphatase;Calcium binding protein | ||||
| Ppp3cb | protein phosphatase 3, catalytic subunit, beta isoform | MGI:107163 | P48453 | X | 9 | 7 | 3 | 2 | 4 | Phosphatase | Protein phosphatase;Calcium binding protein | ||||||||
| Prdx1 | peroxiredoxin 1 | MGI:99523 | P35700 | G00000387 | X | X | X | X | 4 | 7 | 10 | 14 | Enzymes | Peroxidase | |||||
| Prdx2 | peroxiredoxin 2 | MGI:109486 | Q61171 | G00000437 | X | X | X | X | 2 | 2 | 3 | Enzymes | Peroxidase | ||||||
| Prdx3 | peroxiredoxin 3 | MGI:88034 | P20108 | 2 | Enzymes | Peroxidase | |||||||||||||
| Prkar2b | protein kinase, cAMP dependent regulatory, type II beta | MGI:97760 | P31324 | G00000155 | X | X | X | 3 | Kinase | Kinase modulator | |||||||||
| Prrt1 | proline-rich transmembrane protein 1 | MGI:1932118 | O35449 | G00001134 | X | X | X | 2 | 2 | 2 | 3 | Unclassified | Unclassified | ||||||
| Psat1 | phosphoserine aminotransferase 1 | MGI:2183441 | Q99K85 | 3 | Enzymes | Transaminase | |||||||||||||
| Psmb5 | proteasome (prosome, macropain) subunit, beta type 5 | MGI:1194513 | Q91X53 | 2 | Enzymes | Other proteases | |||||||||||||
| Rab6 | RAB6, member RAS oncogene family | MGI:894313 | P35279 | G00001139 | X | X | 4 | G-protein signalling | Small GTPase | ||||||||||
| Rac1 | RAS-related C3 botulinum substrate 1 | MGI:97845 | Q3TLP8 | G00000231 | X | X | X | X | 2 | 2 | 3 | 3 | G-protein signaling | Small GTPase | |||||
| Rap1gds1 | RAP1, GTP-GDP dissociation stimulator 1 | MGI:2385189 | Q921Q5 | 5 | Unclassified | Molecular function unclassified | |||||||||||||
| Rps14 | ribosomal protein S14 | MGI:98107 | P62264 | G00000533 | X | X | X | 2 | 3 | 2 | Transcription/ Translation | Ribosomal protein | |||||||
| Rps3 | ribosomal protein S3 | MGI:1350917 | P62908 | G00000514 | X | X | X | 2 | 2 | 3 | 4 | Transcription/ Translation | Ribosomal protein | ||||||
| Rpsa | ribosomal protein SA | MGI:105381 | P14206 | G00001219 | X | X | 3 | Unclassified | Molecular function unclassified | ||||||||||
| Scrn1 | secernin 1 | MGI:1917188 | Q9CZC8 | 2 | Vesicular/ Trafficking/ Transport | Other miscellaneous function protein | |||||||||||||
| Sdha | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | MGI:1914195 | Q8K2B3 | X | 7 | 6 | 8 | 6 | Enzymes | Dehydrogenase | |||||||||
| Sept11 | septin 11 | MGI:1277214 | Q8C1B7 | G00000221 | X | X | X | 2 | 2 | 2 | 2 | Cytoeskeletal/ Structural/ Cell adhesion | Cytoskeletal protein;Small GTPase | ||||||
| Sept2 | septin 2 | MGI:97298 | Q3U9Y5 | G00000212 | X | X | 3 | G-protein signaling | Cytokinesis | ||||||||||
| Sept5 | septin 5 | MGI:1195461 | Q9Z2Q6 | G00000892 | X | X | X | X | 2 | 2 | 4 | Cytoeskeletal/ Structural/ Cell adhesion | Cytoskeletal protein;Small GTPase | ||||||
| Sf3b3 | splicing factor 3b, subunit 3 | MGI:1289341 | Q921M3 | 2 | Transcription/ Translation | mRNA splicing factor | |||||||||||||
| Sfxn3 | sideroflexin 3 | MGI:2137679 | Q91V61 | G00000734 | X | X | X | X | 3 | 3 | 3 | 4 | 3 | Receptors/ Channels/ Transporters | Cation transporter;Other transfer/carrier protein | ||||
| Sh3gl2 | SH3-domain GRB2-like 2 | MGI:700009 | Q62420 | 3 | Vesicular/ Trafficking/ Transport | Molecular function unclassified | |||||||||||||
| Sh3glb2 | SH3-domain GRB2-like endophilin B2 | MGI:2385131 | Q3U2J4 | 2 | Vesicular/ Trafficking/ Transport | Molecular function unclassified | |||||||||||||
| Shank1 | SH3/ankyrin domain gene 1 | MGI:3613677 | OTTMUSP00000017680 | G00000862 | X | X | X | X | X | x | 4 | Cytoeskeletal/ Structural/ Cell adhesion | Unclassified | ||||||
| Sirt2 | sirtuin 2 (silent mating type information regulation 2, homolog) 2 (S. cerevisiae) | MGI:1927664 | Q8VDQ8 | 3 | Transcription/ Translation | Chromatin/chromatin-binding protein;Deacetylase | |||||||||||||
| Slc12a5 | solute carrier family 12, member 5 | MGI:1862037 | Q80TI5 | G00000115 | X | X | X | 4 | Receptors/ Channels/ Transporters | Cation transporter;Other transporter | |||||||||
| Slc1a2 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | MGI:101931 | P43006 | G00000101 | X | X | X | X | 4 | 3 | 2 | 4 | 4 | Receptors/ Channels/ Transporters | Other transporter | ||||
| Slc1a3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 | MGI:99917 | P56564 | 3 | 3 | 3 | Receptors/ Channels/ Transporters | Other transporter | |||||||||||
| Slc25a12 | solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | MGI:1926080 | Q8BH59 | G00000735 | X | X | X | X | 8 | Receptors/ Channels/ Transporters | Mitochondrial carrier;Calmodulin related | ||||||||
| Slc25a13 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 | MGI:1354721 | Q9QXX4 | G00000748 | X | X | 3 | Receptors/ Channels/ Transporters | Mitochondrial carrier;Calmodulin related | ||||||||||
| Slc25a22 | solute carrier family 25 (mitochondrial carrier, glutamate), member 22 | MGI:1915517 | Q9D6M3 | G00001297 | X | X | X | X | 2 | Receptors/ Channels/ Transporters | Mitochondrial carrier protein | ||||||||
| Slc25a3 | solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 | MGI:1353498 | Q3THU8 | G00000739 | X | X | 3 | Receptors/ Channels/ Transporters | Transporter;Mitochondrial carrier protein | ||||||||||
| Slc25a31 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 | MGI:1920583 | Q3V132 | G00001131 | X | X | X | 3 | Receptors/ Channels/ Transporters | Transporter;Mitochondrial carrier protein | |||||||||
| Slc25a4 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 | MGI:1353495 | P48962 | G00000732 | X | X | X | X | X | 10 | 2 | 3 | 6 | Receptors/ Channels/ Transporters | Transporter;Mitochondrial carrier protein | ||||
| Slc25a5 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 | MGI:1353496 | P51881 | G00000999 | X | X | X | X | X | 9 | 2 | 2 | 3 | 6 | Receptors/ Channels/ Transporters | Transporter;Mitochondrial carrier protein | |||
| Slc2a1 | solute carrier family 2 (facilitated glucose transporter), member 1 | MGI:95755 | P17809 | 2 | 2 | Receptors/ Channels/ Transporters | Carbohydrate transporter | ||||||||||||
| Slc4a4 | solute carrier family 4 (anion exchanger), member 4 | MGI:1927555 | O88343 | G00000118 | X | X | X | X | 2 | 3 | 6 | 4 | Receptors/ Channels/ Transporters | Other transporter | |||||
| Slc9a3r1 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | MGI:1349482 | P70441 | X | 4 | 4 | 3 | 7 | 2 | Unclassified | Unclassified | ||||||||
| Snap25 | synaptosomal-associated protein 25 | MGI:98331 | P60879 | G00001282 | X | X | X | 2 | Vesicular/ Trafficking/ Transport | SNARE protein | |||||||||
| Snca | synuclein, alpha | MGI:1277151 | Q9CXF8 | G00000900 | X | X | 3 | Chaperone/ Protein folding/ Signalling | Chaperone;Membrane traffic protein | ||||||||||
| Sod1 | superoxide dismutase 1, soluble | MGI:98351 | P08228 | G00001208 | 3 | Enzymes | Other oxidoreductase | ||||||||||||
| Spna2 | spectrin alpha 2 | MGI:98386 | P16546 | 11|2 | Cytoeskeletal/ Structural/ Cell adhesion | Molecular function unclassified | |||||||||||||
| Spnb2 | spectrin beta 2 | MGI:98388 | Q62261 | G00000546 | X | X | X | X | X | 2 | 2 | 5 | Cytoeskeletal/ Structural/ Cell adhesion | Non-motor actin binding protein | |||||
| St13 | suppression of tumorigenicity 13 | MGI:1917606 | Q99L47 | 3 | Chaperone/ Protein folding/ Signalling | Chaperone | |||||||||||||
| Stip1 | stress-induced phosphoprotein 1 | MGI:109130 | Q60864 | 9 | Unclassified | Molecular function unclassified | |||||||||||||
| Stx1b | syntaxin 1B | MGI:1930705 | P61264 | G00000716 | X | X | X | X | 3 | 3 | 4 | 3 | 2 | Vesicular/ Trafficking/ Transport | SNARE protein | ||||
| Stxbp1 | syntaxin binding protein 1 | MGI:107363 | O08599 | G00000667 | X | X | X | X | X | X | 5 | 5 | 3 | 4 | 4 | Vesicular/ Trafficking/ Transport | Membrane traffic regulatory protein | ||
| Sucla2 | succinate-Coenzyme A ligase, ADP-forming, beta subunit | MGI:1306775 | Q9Z2I9 | G00000453 | X | X | X | 4 | 2 | 5 | 6 | Enzymes | Synthetase;Ligase | ||||||
| Syn2 | synapsin II | MGI:103020 | Q64332 | G00000660 | X | X | X | 3 | 3 | 4 | Vesicular/ Trafficking/ Transport | Non-motor actin binding;Membrane traffic regulatory | |||||||
| Syngap1 | synaptic Ras GTPase activating protein 1 homolog (rat) | MGI:3039785 | Q9QUH6 | G00000009 | X | X | X | X | X | x | 14 | 21 | 15 | 17 | 38 | G-protein signaling | Unclassified | ||
| Syp | synaptophysin | MGI:98467 | Q62277 | G00001137 | X | X | 2 | Vesicular/ Trafficking/ Transport | Protein phosphatase | ||||||||||
| Syt1 | synaptotagmin I | MGI:99667 | P46096 | G00000662 | X | X | X | X | X | 2 | 5 | 2 | 2 | 2 | Vesicular/ Trafficking/ Transport | Membrane traffic regulatory protein | |||
| Tagln3 | transgelin 3 | MGI:1926784 | Q9R1Q8 | G00000597 | X | X | 4 | Cytoeskeletal/ Structural/ Cell adhesion | Non-motor actin binding protein | ||||||||||
| Tkt | transketolase | MGI:105992 | Q545A1 | G00000388 | X | X | 4 | Enzymes | Tyrosine protein kinase receptor;Protein kinase | ||||||||||
| Tmod2 | tropomodulin 2 | MGI:1355335 | Q9JKK7 | G00000752 | X | X | X | 2 | Cytoeskeletal/ Structural/ Cell adhesion | Actin binding cytoskeletal protein | |||||||||
| Tnr | tenascin R | MGI:99516 | Q8BYI9 | G00000562 | X | X | 6 | Cytoeskeletal/ Structural/ Cell adhesion | Cell adhesion;Extracellular matrix glycoprotein | ||||||||||
| Tpm3 | tropomyosin 3, gamma | MGI:1890149 | Q58E70 | G00001001 | X | X | X | 6 | Cytoeskeletal/ Structural/ Cell adhesion | Actin binding motor protein | |||||||||
| Traf3 | TNF receptor-associated factor 3 | MGI:108041 | Q3UHJ1 | G00000131 | X | X | X | 3 | 3 | Signal transduction | Signaling molecule;Miscellaneous function | ||||||||
| Tuba1a* | tubulin, beta polypeptide | MGI:98869 | P05213 | G00001054 | X | X | X | X | X | X | 11 | 14 | 20 | 20 | Cytoeskeletal/ Structural/ Cell adhesion | Tubulin | |||
| Tubb2b* | tubulin, beta 2b | MGI:1920960 | Q7TMM9 | X | 19 | 21 | 24 | 27 | Cytoeskeletal/ Structural/ Cell adhesion | Tubulin | |||||||||
| Tubb6 | tubulin, beta 6 | MGI:1915201 | Q922F4 | G00000807 | X | X | X | X | 12 | 8 | 8 | 13 | 13 | Cytoeskeletal/ Structural/ Cell adhesion | Tubulin | ||||
| Tufm | Tu translation elongation factor, mitochondrial | MGI:1923686 | Q497E7 | 2 | Transcription/ Translation | Translation elongation factor | |||||||||||||
| Txn1 | thioredoxin 1 | MGI:98874 | P10639 | 2 | Enzymes | Other oxidoreductase | |||||||||||||
| Txnl1 | thioredoxin-like 1 | MGI:1860078 | Q3TI92 | 2 | Enzymes | Other oxidoreductase | |||||||||||||
| Uba1 | ubiquitin-like modifier activating enzyme 1 | MGI:98890 | Q542H8 | 11 | Enzymes | Other ligase | |||||||||||||
| Uba52* | ubiquitin A-52 residue ribosomal p. fusion product 1 | MGI:98887 | Q66JP1 | G00001293 | X | X | X | 3 | 3 | 5 | 6 | Transcription/ Translation | Ribosomal protein | ||||||
| Ubb | ubiquitin B | MGI:98888 | Q6NZC5 | G00001292 | X | X | 2 | Signal transduction | Ribosomal protein | ||||||||||
| Ube2n | ubiquitin-conjugating enzyme E2N | MGI:1934835 | P61089 | 2 | Enzymes | Other ligase | |||||||||||||
| Ube2v1 | ubiquitin-conjugating enzyme E2 variant 1 | MGI:1913839 | Q9CZY3 | G00000438 | X | X | 2 | 2 | 5 | Enzymes | Other ligase | ||||||||
| Ube2v2 | ubiquitin-conjugating enzyme E2 variant 2 | MGI:1917870 | Q9D2M8 | 2 | 4 | Enzymes | Other ligase | ||||||||||||
| Uqcrc1 | ubiquinol-cytochrome c reductase core protein 1 | MGI:107876 | Q9CZ13 | G00000948 | X | X | 7 | Enzymes | Reductase;Metalloprotease | ||||||||||
| Uqcrc2 | ubiquinol cytochrome c reductase core protein 2 | MGI:1914253 | Q9CVK7 | G00000938 | X | X | 9 | Enzymes | Reductase;Metalloprotease | ||||||||||
| Vamp2* | synaptobrevin 2, 3 | MGI:1313277 | P63024 | G00000725 | X | X | X | X | X | 2 | 4 | 2 | Vesicular/ Trafficking/ Transport | SNARE protein | |||||
| Vdac1 | voltage-dependent anion channel 1 | MGI:106919 | Q60932 | G00000091 | X | X | X | X | X | 9 | 4 | 4 | 5 | 5 | Receptors/ Channels/ Transporters | Anion channel;Voltage-gated ion channel | |||
| Vdac2 | voltage-dependent anion channel 2 | MGI:106915 | Q60930 | G00000092 | X | X | X | X | X | 4 | 4 | 2 | 3 | 4 | Receptors/ Channels/ Transporters | Anion channel;Voltage-gated ion channel | |||
| Vdac3 | voltage-dependent anion channel 3 | MGI:106922 | Q60931 | G00000858 | X | X | X | 4 | 2 | 5 | Receptors/ Channels/ Transporters | Anion channel;Voltage-gated ion channel | |||||||
| Vsnl1 | visinin-like 1 | MGI:1349453 | Q3TVC0 | 3 | Signal transduction | Calmodulin related protein | |||||||||||||
| Ywhae | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | MGI:894689 | P62259 | G00000148 | X | X | X | X | 3 | 2 | 2 | Signal transduction | Other chaperones | ||||||
Genes marked with an asterisk represent genes whose peptides are common to other genes:
- Cpne5*
- Cpne4, Cpne8.
- Eif1a1*
- Eif1a2.
- Tuba1a*
- Tuba1b, Tuba4c, Tuba1b.
- Hist1h2bj*
- Hist1h2bm Hist1h2be Hist1h2bn Hist1h2bg Hist1h2bp Hist1h2bh Hist1h2bf Hist1h2bb Hist3h2bb Hist1h2bc Hist1h2bl Hist2h2bb.
- Tubb2b*
- Tubb5, Tubb2a, Tubb2c, Tubb4.
- Uba52*
- Ubc, Ubb.
- Vamp2*
- Vamp3.
Supplementary Table 2. Proteins identified by LC-MS/MS in three tandem purifications from wild type mice.
Gene symbols, UniProt and IPI accession numbers and number of approved peptides for each protein identified by:
- LC-MS/MS analysis in the 4 tandem purifications from PSD-95TAP/TAP mice (T1, T2, T3, T4) and 3 tandem
- purifications from wt mice (WT1, WT2, WT3) are shown.
| Number of peptides | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MGI symbol | Protein Acc | IPI Acc | Description | T1 | T2 | T3 | T4 | WT1 | WT2 | WT3 |
| Atp1a2 | Q6ZQ49 | IPI00762871 | IPI:IPI00762871.2|TREMBL:Q6ZQ49|ENSEMBL:ENSMUSP00000095072|VEGA:OTTMUSP00000024454 Tax_Id=10090 Gene_Symbol=Atp1a2 MKIAA0778 protein | 4 | 4 | 8 | 7 | 4 | 2 | 3 |
| Hspa2 | P17156 | IPI00331546 | IPI:IPI00331546.2|SWISS-PROT:P17156 Tax_Id=10090 Gene_Symbol=Hspa2 Heat shock-related 70 kDa protein 2 | 7 | 7 | 7 | 9 | 3 | 1 | 1 |
| Glud1 | XP_992848 | IPI00753095 | IPI:IPI00753095.1|REFSEQ:XP_992848 Tax_Id=10090 Gene_Symbol=Glud1 similar to Glutamate dehydrogenase 1, mitochondrial precursor (GDH) isoform 2 | 8 | 3 | 7 | 7 | 6 | 3 | 4 |
| Atp1a3 | Q6PIC6 | IPI00122048 | IPI:IPI00122048.2|SWISS-PROT:Q6PIC6|TREMBL:Q8CGD9;Q8R0B0;Q8R0E8|VEGA:OTTMUSP00000017754 Tax_Id=10090 Gene_Symbol=Atp1a3 Sodium/potassium-transporting ATPase alpha-3 chain | 9 | 8 | 11 | 9 | 6 | 2 | 4 |
| Aldoa | P05064 | IPI00221402 | IPI:IPI00221402.7|SWISS-PROT:P05064|TREMBL:Q5FWB7;Q6NY00|ENSEMBL:ENSMUSP00000032934|REFSEQ:NP_031464 Tax_Id=10090 Gene_Symbol=Aldoa Fructose-bisphosphate aldolase A | 13 | 7 | 10 | 11 | 6 | - | 3 |
| Hspa8 | P63017 | IPI00323357 | IPI:IPI00323357.3|SWISS-PROT:P63017|TREMBL:Q3KQJ4;Q3TB63;Q3TEK2;Q3TF16;Q3TH04;Q3TH56;Q3TQ13;Q3TRH3;Q3TZJ3;Q3U9L2;Q3UBA6;Q3UBR0;Q3UDS0;Q3ULM1;Q3UXU1;Q504P4;Q9QZ38|ENSEMBL:ENSMUSP00000015800|REFSEQ:NP_112442 Tax_Id=10090 Gene_Symbol=Hspa8 Heat shock cognate | 21 | 22 | 19 | 23 | 5 | 2 | 3 |
| Atp1a1 | Q8VDN2 | IPI00311682 | IPI:IPI00311682.5|SWISS-PROT:Q8VDN2|TREMBL:Q3TXF9;Q3UCH8;Q3UF49;Q8K2R2;Q8K2Y5;Q8R3M4|ENSEMBL:ENSMUSP00000039657;ENSMUSP00000062842|REFSEQ:NP_659149|VEGA:OTTMUSP00000017686 Tax_Id=10090 Gene_Symbol=Atp1a1 Sodium/potassium-transporting ATPase alpha-1 chain | 6 | 4 | 7 | 6 | 4 | 1 | 3 |
| - | ENSMUSP00000097709 | IPI00761732 | IPI:IPI00761732.1|ENSEMBL:ENSMUSP00000097709 Tax_Id=10090 Gene_Symbol=- 49 kDa protein | 2 | 2 | 8 | 3 | 3 | 1 | 1 |
| Eef1a1 | P10126 | IPI00307837 | IPI:IPI00307837.5|SWISS-PROT:P10126|TREMBL:Q3TII3;Q3UA81;Q3UZQ3;Q4FZE5;Q58E64|ENSEMBL:ENSMUSP00000042457|REFSEQ:NP_034236|VEGA:OTTMUSP00000021291 Tax_Id=10090 Gene_Symbol=Eef1a1 Elongation factor 1-alpha 1 | 2 | 2 | 7 | 4 | 3 | 1 | 1 |
| Eef1a2 | P62631 | IPI00119667 | IPI:IPI00119667.1|SWISS-PROT:P62631|ENSEMBL:ENSMUSP00000054556|REFSEQ:NP_031932|VEGA:OTTMUSP00000017863 Tax_Id=10090 Gene_Symbol=Eef1a2 Elongation factor 1-alpha 2 | 2 | 2 | 7 | 3 | 3 | 1 | 1 |
| Crmp1 | P97427 | IPI00312527 | IPI:IPI00312527.4|SWISS-PROT:P97427|TREMBL:Q3TXY0;Q6P1J1|ENSEMBL:ENSMUSP00000031004|REFSEQ:NP_031791 Tax_Id=10090 Gene_Symbol=Crmp1 Crmp1 protein | 3 | - | 1 | 8 | 3 | - | - |
| Rpsa | P14206 | IPI00123604 | IPI:IPI00123604.4|SWISS-PROT:P14206|ENSEMBL:ENSMUSP00000035105|REFSEQ:NP_035159 Tax_Id=10090 Gene_Symbol=Rpsa 40S ribosomal protein SA | - | - | - | - | 3 | - | - |
| Atp1a4 | Q9WV27 | IPI00378485 | IPI:IPI00378485.2|SWISS-PROT:Q9WV27|TREMBL:Q91ZN9|ENSEMBL:ENSMUSP00000007346|VEGA:OTTMUSP00000024456 Tax_Id=10090 Gene_Symbol=Atp1a4 Sodium/potassium-transporting ATPase alpha-4 chain | - | 2 | - | - | 2 | 1 | 1 |
| Ckb | Q04447 | IPI00136703 | IPI:IPI00136703.1|SWISS-PROT:Q04447|ENSEMBL:ENSMUSP00000001304|REFSEQ:NP_067248 Tax_Id=10090 Gene_Symbol=Ckb Creatine kinase B-type | 3 | 2 | 3 | 4 | 1 | 2 | 3 |
| Prkcb | P68404 | IPI00227898 | IPI:IPI00227898.3|SWISS-PROT:P68404|TREMBL:A0JNZ5|ENSEMBL:ENSMUSP00000070019|REFSEQ:NP_032881|VEGA:OTTMUSP00000017803 Tax_Id=10090 Gene_Symbol=Prkcb1 Protein kinase C beta type | - | 3 | 2 | 3 | - | 4 | 5 |
| Cap2 | Q9CYT6 | IPI00112001 | IPI:IPI00112001.1|SWISS-PROT:Q9CYT6|TREMBL:Q8BKZ3|ENSEMBL:ENSMUSP00000021802|REFSEQ:NP_080332 Tax_Id=10090 Gene_Symbol=Cap2 Adenylyl cyclase-associated protein 2 | - | - | - | 1 | - | 1 | 7 |
| Ppm1b | P36993-5 | IPI00222989 | IPI:IPI00222989.1|SWISS-PROT:P36993-5 Tax_Id=10090 Gene_Symbol=Ppm1b Isoform Beta-5 of Protein phosphatase 1B | - | - | 1 | 2 | - | 1 | 4 |
| Pkm2 | P52480-2 | IPI00845840 | IPI:IPI00845840.1|SWISS-PROT:P52480-2 Tax_Id=10090 Gene_Symbol=Pkm2 Isoform M1 of Pyruvate kinase isozymes M1/M2 | 7 | 6 | 11 | 12 | - | 1 | 2 |
| - | Q52L64 | IPI00785509 | IPI:IPI00785509.1|TREMBL:Q52L64|ENSEMBL:ENSMUSP00000100179 Tax_Id=10090 Gene_Symbol=- Hypothetical protein | - | 1 | - | 4 | - | - | 4 |
| Prkca | Q3TQ39 | IPI00652220 | "IPI:IPI00652220.1|TREMBL:Q3TQ39|ENSEMBL:ENSMUSP00000097875|VEGA:OTTMUSP00000017462 Tax_Id=10090 Gene_Symbol=Prkca 0 day neonate cerebellum cDNA, RIKEN full-length enriched library, clone:C230094C20 product:similar to Protein kinase C beta 2" | - | 2 | - | - | - | - | 3 |
Supplementary Table 3. List of known PSD-95 primary interactors extracted from UniHI database and manually curated.
The HGNC symbols and number of peptides obtained in the single step and four tandem replicates are shown. The PubMed ID and the method used to see the interaction with PSD-95 are also shown.
| Number of Peptides | ||||||||
|---|---|---|---|---|---|---|---|---|
| HGNC symbol | Single | T1 | T2 | T3 | T4 | Interaction Database | PubMed ID | Determination Method |
| ADAM22 | 4 | 12 | 16 | 24 | 31 | HPRD-Binary|BioGrid | 18206289|16990550 | In vivo|In vitro |
| ATP2B2 | 6 | BIND|HPRD-Binary|BioGrid | 11274188 | In vitro | ||||
| ATP2B4 | 5 | HPRD-Binary|BioGrid | 11274188 | In vitro | ||||
| BAIP2 | 14 | 20 | 12 | 13 | 13 | HPRD-Binary|BioGrid | 11937501 | In vivo|In vitro |
| BEGAIN | 4 | 8 | 12 | 16 | HPRD-Binary|HPRD-Complex|BioGrid | 9756850 | Y2H|In vitro|In vivo | |
| CACNG2 | 2 | 2 | 2 | 3 | HPRD-Binary|BioGrid | 12359873|11805122 | Y2H|In vitro|In vivo | |
| CAMKIIa | 7|2 | 11 | 3 | 4 | 11 | manually curated | 17156196 | In vitro|In vivo |
| CIT | 11 | HPRD-Binary|BioGrid | 9870942 | Y2H|In vivo | ||||
| DLG1 | 17 | 24 | 42 | 34 | 42 | HPRD-Binary|BioGrid | 16332687|17855605 | Y2H|In vivo |
| DLG2 | 42 | 49 | 67 | 69 | 80 | HPRD-Binary|BioGrid | 9182804|17855605 | In vivo |
| DLG3 | 16 | 27 | 17 | 14 | 22 | HPRD-Binary|HPRD-Complex|BioGrid | 10026200|9808460 | Y2H|In vivo|In vitro |
| DLGAP1 | 10 | 11 | 9 | 14 | 24 | HOMOMINT|HPRD-Binary|HPRD-Complex|BioGrid | 16351748|9115257|9024696|9286858|10844022 | Computational prediction|Y2H|In vivo|In vitro |
| DLGAP2 | 9 | 18 | 12 | 19 | 24 | HPRD-Binary|BioGrid | 9286858|9115257 | In vitro|Y2H|In vivo |
| DLGAP3 | 3 | 16 | 8 | 7 | 15 | BioGrid | 9115257 | In vitro |
| DLGAP4 | 12 | 14 | 9 | 7 | 14 | HPRD-Binary|BioGrid | 9115257 | Y2H|In vitro |
| DYNLL1 | 5 | HPRD-Binary|BioGrid | 14760703 | In vitro | ||||
| GDA | 29|4 | 29 | 26 | 29 | 31 | HPRD-Binary|BioGrid | 10595517 | In vitro|Y2H|In vivo |
| GRIK2 | 2 | 4 | 6 | HPRD-Binary|BioGrid | 11276111|12597860|9808460|11744724 | In vitro|In vivo | ||
| GRIK5 | 2 | 3 | 7 | HPRD-Complex|HPRD-Binary|BioGrid | 9808460|11279111 | In vivo|In vitro | ||
| GRIN1 | 31 | 29 | 40 | 50 | 55 | HOMOMINT|HPRD-Binary | 16351748|15030493|12068077|11506858|9009191|14732708| | Computational prediction|In vivo|In vitro |
| GRIN2A | 15 | 24 | 31 | 36 | 46 | HOMOMINT|HPRD-Binary|BioGrid | 16351748|11937501|12419528|7569905|11222640|10336672 | Computational prediction|In vitro|In vivo|Y2H |
| GRIN2B | 32 | 44 | 54 | 67 | 78 | HOMOMINT|HPRD-Binary|HPRD-Complex|BioGrid | 16351748|9581762|7569905|10488080|8601796|11937501|11029657 | Computational prediction|Y2H|In vitro|In vivo |
| GRIN2C | 5 | HPRD-Binary|BioGrid | 11937501|7569905 | In vitro|Y2H|In vivo | ||||
| GRIN2D | 2 | 3 | 6 | 9 | 10 | HPRD-Binary|BioGrid | 7569905 | Y2H|In vitro|In vivo |
| KCNA1 | 2 | 6 | 5 | 5 | 6 | HPRD-Binary|BioGrid | 12435606|8938729|7477295 | Y2H|In vitro |
| KCNA2 | 3 | 4 | 5 | 7 | 6 | HPRD-Binary|BioGrid | 7477295|8938729|12435606 | In vitro|Y2H |
| KCNA3 | 1 | 3 | 4 | 6 | 5 | HPRD-Binary | 8938729|12435606 | In vitro|Y2H |
| KCNA4 | 2 | 3 | 5 | 5 | HOMOMINT|HPRD-Binary|BioGrid | 16351748|11723117|11744724|9581762|7477295|8601796|8938729 | Computational prediction|In vivo|In vitro|Y2H | |
| KCNJ10 | 5 | 3 | 3 | 4 | 6 | HPRD-Binary|BioGrid | 9148889 | In vitro|Y2H|In vivo |
| KCNJ4 | 1 | 4 | 6 | 6 | 8 | HPRD-Binary|BioGrid | 10627592 | Y2H|In vivo |
| LRP1 | 2 | 3 | HPRD-Binary|BioGrid | 10827173|10880444 | In vitro|Y2H|In vivo | |||
| NLGN2 | 2 | HPRD-Binary|BioGrid | 9278515 | Y2H|In vitro | ||||
| NLGN3 | 2 | HPRD-Binary|BioGrid | 9278515 | Y2H|In vitro | ||||
| SHANK1 | 4 | HPRD-Binary|BioGrid | 10433268 | In vitro|In vivo | ||||
| SYNGAP1 | 14 | 21 | 15 | 17 | 38 | HPRD-Binary|BioGrid | 9581761 | In vitro|In vivo|Y2H |
Supplementary Table 4. EmPAI values from single step and tandem purifications.
EmPAI values for each protein present in both single step and tandem purifications were normalised to the total protein emPAI in each set and then normalised to the emPAI value of the bait protein (PSD-95).
| UniProt Acc | MGI Symbol | Normalised emPAI (single step) x 100 | Normalised emPAI (tandem) x 100 | Ratio emPAI (tandem/single step) |
|---|---|---|---|---|
| Q9R1V6 | Adam22 | 0.13 | 0.45 | 3.45 |
| Q6PFD5 | Dlgap3 | 0.09 | 0.21 | 2.33 |
| Q03391 | Grin2d | 0.06 | 0.11 | 2.05 |
| Q9Z2W9 | Gria3 | 0.12 | 0.24 | 1.96 |
| Q8R0S2 | Iqsec1 | 0.08 | 0.16 | 1.94 |
| Q5DU25 | Iqsec2 | 0.22 | 0.41 | 1.90 |
| Q8BZM2 | Anks1b | 0.32 | 0.55 | 1.72 |
| Q7TNS5 | B630019K06Rik | 0.43 | 0.73 | 1.70 |
| Q3UXZ6 | Fam81a | 0.31 | 0.53 | 1.70 |
| P16388 | Kcna1 | 0.15 | 0.26 | 1.68 |
| P23819 | Gria2 | 0.14 | 0.22 | 1.58 |
| Q3UP61 | Dlg1 | 0.79 | 1.18 | 1.49 |
| P35436 | Grin2a | 0.36 | 0.53 | 1.47 |
| Q9Z307 | Kcnj16 | 0.24 | 0.35 | 1.45 |
| Q01097 | Grin2b | 0.90 | 1.20 | 1.33 |
| P15105 | Glul | 0.77 | 1.03 | 1.33 |
| Q9WV31 | Arc | 0.81 | 1.04 | 1.28 |
| P62482 | Kcnab2 | 0.37 | 0.47 | 1.25 |
| O88343 | Slc4a4 | 0.08 | 0.10 | 1.25 |
| Q8BJ42 | Dlgap2 | 0.30 | 0.37 | 1.25 |
| Q91XM9 | Dlg2 | 2.36 | 2.85 | 1.21 |
| P63141 | Kcna2 | 0.29 | 0.33 | 1.11 |
| P60202 | Plp1 | 0.44 | 0.48 | 1.08 |
| Q60931 | Vdac3 | 0.42 | 0.45 | 1.08 |
| Q9CZY3 | Ube2v1 | 0.32 | 0.34 | 1.06 |
| Q62108 | Dlg4 | 3.30 | 3.30 | 1.00 |
| Q9JIA1 | Lgi1 | 0.46 | 0.46 | 0.99 |
| Q9QUH6 | Syngap1 | 0.40 | 0.40 | 0.98 |
| P35486 | Pdha1 | 0.20 | 0.19 | 0.97 |
| Q9R0P5 | Dstn | 0.40 | 0.38 | 0.95 |
| A2AI21 | Grin1 | 1.19 | 1.06 | 0.89 |
| O54983 | Crym | 0.29 | 0.25 | 0.88 |
| Q61990 | Pcbp2 | 0.20 | 0.17 | 0.87 |
| P62908 | Rps3 | 0.18 | 0.15 | 0.87 |
| Q9D415 | Dlgap1 | 0.35 | 0.30 | 0.85 |
| P46096 | Syt1 | 0.14 | 0.11 | 0.78 |
| O89112 | Lancl1 | 0.24 | 0.19 | 0.77 |
| Q52KF7 | Dlg3 | 0.67 | 0.49 | 0.74 |
| P60335 | Pcbp1 | 0.29 | 0.21 | 0.70 |
| P18872 | Gnao1 | 0.94 | 0.63 | 0.67 |
| Q64332 | Syn2 | 0.21 | 0.14 | 0.66 |
| P16858 | Gapdh | 0.88 | 0.58 | 0.65 |
| Q91VR2 | Atp5c1 | 0.32 | 0.20 | 0.63 |
| Q91V61 | Sfxn3 | 0.27 | 0.16 | 0.61 |
| Q8BKX1 | Baiap2 | 0.76 | 0.45 | 0.59 |
| P11798 | Camk2a | 0.51 | 0.30 | 0.58 |
| P62874 | Gnb1 | 0.32 | 0.18 | 0.56 |
| P70441 | Slc9a3r1 | 0.28 | 0.16 | 0.56 |
| P56564 | Slc1a3 | 0.24 | 0.14 | 0.56 |
| P61264 | Stx1b | 0.29 | 0.16 | 0.56 |
| Q9R111 | Gda | 2.88 | 1.60 | 0.55 |
| Q5SVI3 | Camk2b | 0.49 | 0.27 | 0.54 |
| P63085 | Mapk1 | 0.39 | 0.21 | 0.53 |
| B1AZP2 | Dlgap4 | 0.42 | 0.21 | 0.51 |
| Q9D051 | Pdhb | 0.54 | 0.27 | 0.49 |
| Q8QZT1 | Acat1 | 0.21 | 0.11 | 0.49 |
| Q922F4 | Tubb6 | 1.15 | 0.55 | 0.47 |
| P43006 | Slc1a2 | 0.26 | 0.12 | 0.45 |
| P52480 | Pkm2 | 0.59 | 0.26 | 0.45 |
| Q60930 | Vdac2 | 0.45 | 0.20 | 0.45 |
| O08599 | Stxbp1 | 0.22 | 0.10 | 0.44 |
| Q9JM63 | Kcnj10 | 0.65 | 0.28 | 0.43 |
| P16330 | Cnp | 0.75 | 0.31 | 0.41 |
| P51863 | Atp6v0d1 | 0.40 | 0.17 | 0.41 |
| A2A817 | Park7 | 2.10 | 0.79 | 0.37 |
| Q8K0U4 | Hspa12a | 0.12 | 0.04 | 0.37 |
| P47754 | Capza2 | 0.30 | 0.11 | 0.36 |
| Q91V12 | Acot7 | 0.45 | 0.16 | 0.35 |
| Q61553 | Fscn1 | 0.21 | 0.07 | 0.32 |
| P08551 | Nefl | 0.30 | 0.09 | 0.31 |
| P09411 | Pgk1 | 0.77 | 0.23 | 0.31 |
| P63328 | Ppp3ca | 1.10 | 0.33 | 0.30 |
| Q60932 | Vdac1 | 0.99 | 0.25 | 0.25 |
| P48453 | Ppp3cb | 0.60 | 0.14 | 0.23 |
| Q6GQW0 | Btbd11 | 0.27 | 0.06 | 0.22 |
| P10126 | Eef1a1 | 0.41 | 0.09 | 0.21 |
| Q99KI0 | Aco2 | 0.29 | 0.06 | 0.20 |
| P46460 | Nsf | 0.61 | 0.12 | 0.19 |
| Q5SXR6 | Cltc | 0.47 | 0.09 | 0.19 |
| P51881 | Slc25a5 | 0.73 | 0.13 | 0.18 |
| P48962 | Slc25a4 | 0.85 | 0.15 | 0.18 |
| P14094 | Atp1b1 | 1.29 | 0.22 | 0.17 |
| Q05CJ5 | Atp2b1 | 1.30 | 0.23 | 0.17 |
Supplementary Table 5. Comparison to proteins found in receptor complexes and synaptic lists.
Numbers of common proteins found in the MASC/NRC, AMPA receptor (AMPA), metabotrobic glutamate receptor 5 (mGluR5), PSD and PSP lists (Collins et al., 2006) and PSD-95 immunoprecipitation list (Dosemeci et al., 2007) with all the proteins indentified in this study (301 proteins, Supplementary Table 1) and in the PSD-95 core complexes (118 proteins).
| Synaptic Complex | PSD-95 Complex | PSD-95 Core Complex |
|---|---|---|
| MASC/NRC | 69 | 48 |
| AMPA receptor | 7 | 7 |
| mGLUR5 | 17 | 9 |
| PSD | 183 | 89 |
| PSP | 188 | 90 |
| Dosemeci et al. | 79 | 49 |
| Total Proteins | 301 | 118 |
Supplementary Table 6. Protein nodes of the interaction network's MCC.
Protein nodes are ranked by average shortest path (ASP) calculated as the average number of shortest paths between the node and all the other nodes in the network.
| Protein Node | ASP |
|---|---|
| Dlg4 | 1.325 |
| Dlg1 | 1.7 |
| Dlg3 | 1.75 |
| Grin1 | 1.775 |
| Camk2a | 1.8 |
| Dlg2 | 1.825 |
| Grin2a | 1.875 |
| Grin2b | 1.875 |
| Syngap1 | 1.95 |
| Dlgap1 | 1.975 |
| Cacng2 | 1.975 |
| Grik2 | 2 |
| Dlgap4 | 2 |
| Grin2d | 2.025 |
| Kcna4 | 2.025 |
| Kcna2 | 2.1 |
| Kcnj4 | 2.15 |
| Grik5 | 2.15 |
| Cypin | 2.15 |
| Kcnj10 | 2.175 |
| Kcna1 | 2.2 |
| Kcna3 | 2.2 |
| Dlgap2 | 2.225 |
| Adam22 | 2.225 |
| Baiap2 | 2.225 |
| Gria1 | 2.25 |
| Begain | 2.275 |
| Dlgap3 | 2.275 |
| Camk2b | 2.475 |
| Kcnab1 | 2.5 |
| Nefl | 2.575 |
| Spnb2 | 2.6 |
| Gria2 | 2.7 |
| Cltc | 2.7 |
| Pppp3ca | 2.725 |
| Gria4 | 2.775 |
| Kcnab2 | 3.05 |
| Rac1 | 3.175 |
| Gria3 | 3.175 |
| Lgi1 | 3.175 |
| Average ASP | 2.2525 |
Supplementary Table 7. Gene targeting and genotyping primers.
Primers used for the gene targeting vector generation and genotyping screening.
| Primers | Sequence |
|---|---|
| XbaIHATF1 | 5'-CGGCGCTCTAGAAAGGATCATCTCATCCACAATGTCC |
| HATR1 | 5'TGCTGCTGCCGTTGGGAGCTCACCCCTGAAAATACAAATTCTCGATCTTGTTGTGGGGCATGAGCG |
| F1Flag | 5'-GAGAATTTGTATTTTCAGGGTGAGCTCCCAACGGCAGCAGACTACAAAGACCATGACGGTG |
| BclIFlagR1 | 5'-GTCGGTACTTATCGTCTCTTGTCATACACCGTTGATCAC |
| Psd95HAXhoIF | 5'-GCCACATCTGTAACTCCGCTCGAGGCCGTGAG |
| Psd95HAXbaIR | 5'CTGCTCTAGACCGAGTCTCTCTCGGGCTGGGA |
| Psd95HAAcc65IF | 5'-GCCTGGCTTGGCCTG'CTACGGGGTACCTTCCTGC |
| Psd95HABglIIR | 5'-GACCTAGATCTGCGGCCGCACGGGGGGACCGGTGGGAGGTGTGTGAAAG |
| Psd95HANdeIF | 5'-GGGTAATCTCCAGGCAAGAGTTTTATGCCCCACTTCCCCTTTCCT |
| Psd95HAPmlIR | 5'-GAGAGAGCCAGAGCCAGGCTCAGCCAAGTGCCGGCGTACAC |
| Psd95HAAscIF | 5'CATTTGGCGCGCCGGGGAAGTCGGACATGCCATCCAAC |
| Psd95HAPmeIR | 5'-CGTCACAAGCATCTGTAATCCTAGCTTTGTTTAAACCCCAGC5' |
| Psd95F5 | 5'-GGTCCCAGCCCGAGAGAGACTC |
| pneoR4 | 5'-CGTCCTGCACGACGCGAGCTGC |
| pneoF3 | 5'-GCCTCTGAGCTATTCCAGAAGTAG |
| Psd95R3 | 5'-CCTTGGTGTATGCACACATGC |
| Psd95R6 | 5'-GGCGGCAGCATTTCCTGTCCTC |
